IlGFInsulin / insulin-like growth factor / relaxin family. |
|---|
| SMART accession number: | SM00078 |
|---|---|
| Description: | Family of proteins including insulin, relaxin, and IGFs. Insulin decreases blood glucose concentration. |
| Interpro abstract (IPR016179): | The insulin family of proteins groups together several evolutionarily related active peptides [ (PUBMED:6107857) ]: these include insulin [ (PUBMED:6243748) (PUBMED:503234) ], relaxin [ (PUBMED:10601981) (PUBMED:8735594) ], insect prothoracicotropic hormone (bombyxin) [ (PUBMED:8683595) ], insulin-like growth factors (IGF1 and IGF2) [ (PUBMED:2036417) (PUBMED:1319992) ], mammalian Leydig cell-specific insulin-like peptide (gene INSL3), early placenta insulin-like peptide (ELIP) (gene INSL4), locust insulin-related peptide (LIRP), molluscan insulin-related peptides (MIP), and Caenorhabditis elegans insulin-like peptides. The 3D structures of a number of family members have been determined [ (PUBMED:2036417) (PUBMED:1319992) (PUBMED:9141131) ]. The fold comprises two polypeptide chains (A and B) linked by two disulphide bonds: all share a conserved arrangement of 4 cysteines in their A chain, the first of which is linked by a disulphide bond to the third, while the second and fourth are linked by interchain disulphide bonds to cysteines in the B chain. Insulin is found in many animals, and is involved in the regulation of normal glucose homeostasis. It also has other specific physiological effects, such as increasing the permeability of cells to monosaccharides, amino acids and fatty acids, and accelerating glycolysis and glycogen synthesis in the liver [ (PUBMED:6243748) ]. Insulin exerts its effects by interaction with a cell-surface receptor, which may also result in the promotion of cell growth [ (PUBMED:6243748) ]. Insulin is synthesised as a prepropeptide from which an endoplasmic reticulum-targeting sequence is cleaved to yield proinsulin. The sequence of prosinsulin contains 2 well-conserved regions (designated A and B), separated by an intervening connecting region (C), which is variable between species [ (PUBMED:503234) ]. The connecting region is cleaved, liberating the active protein, which contains the A and B chains, held together by 2 disulphide bonds [ (PUBMED:503234) ]. This entry represents a disulphide-rich alpha-helical domain found in insulin [ (PUBMED:12595704) ], as well as in related proteins, such as insulin-like growth factor [ (PUBMED:15642270) ], relaxin [ (PUBMED:1656049) ] and bombyxin [ (PUBMED:7473749) ]. |
| GO component: | extracellular region (GO:0005576) |
| GO function: | hormone activity (GO:0005179) |
| Family alignment: |
There are 4123 IlGF domains in 4108 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing IlGF domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with IlGF domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing IlGF domain in the selected taxonomic class.
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Schwabe C, Bullesbach EE
- Relaxin: structures, functions, promises, and nonevolution.
- FASEB J. 1994; 8: 1152-60
- Display abstract
During the last two decades synthetic chemistry and molecular biology have transformed the little-known parturition-mediating factor relaxin into a chemically defined entity. Relaxin is a disulfide bond analog of insulin that shows no cross-reactivity to the insulin receptors, causes widening of the birth canal in most mammals, and has additional or different functions in various species. The receptor interaction site in relaxin has been located quite precisely in the midregion of the B chain helix and is now known to involve two arginine residues that project from the alpha helix in an n, n + 4 configuration. The A chain of relaxin which appears to be uninvolved in receptor binding, is nonetheless essential. In fact, a major structural determinant in relaxin and insulin responsible for achieving either an insulin-like or a relaxin-like conformation is located in the penultimate position in the intrachain loop of the A chains.
- Murray-Rust J, McLeod AN, Blundell TL, Wood SP
- Structure and evolution of insulins: implications for receptor binding.
- Bioessays. 1992; 14: 325-31
- Display abstract
Insulin is a member of a family of hormones, growth factors and neuropeptides which are found in both vertebrates and invertebrates. A common 'insulin fold' is probably adopted by all family members. Although the specificities of receptor binding are different, there is a possibility of co-evolution of polypeptides and their receptors.
- Blundell TL, Humbel RE
- Hormone families: pancreatic hormones and homologous growth factors.
- Nature. 1980; 287: 781-7
- Display abstract
The growing realization that biologically active polypeptides can be grouped in families, the members of which show structural and functional relatedness, is illustrated by the four families which are represented in the pancreas by the hormones insulin, glucagon, somatostatin and pancreatic polypeptide.
- Bedarkar S, Turnell WG, Blundell TL, Schwabe C
- Relaxin has conformational homology with insulin.
- Nature. 1977; 270: 449-51
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the IlGF domain.
Protein Disease Insulin (P01308) (SMART) OMIM:176730: Diabetes mellitus, rare form ; MODY, one form ; Hyperproinsulinemia, familial - Metabolism (metabolic pathways involving proteins which contain this domain)
-
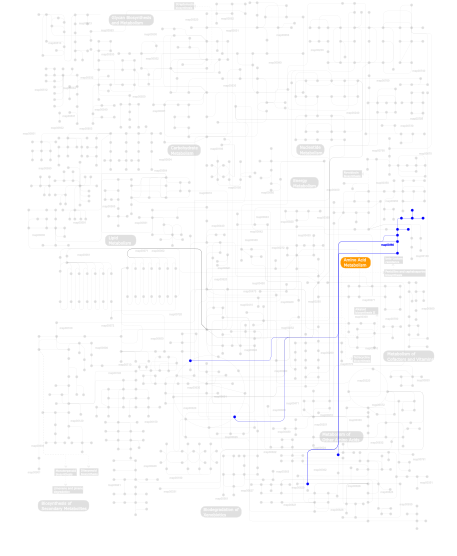
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 10.75 map05215 Prostate cancer 10.75 map04150 mTOR signaling pathway 10.75 map04914 Progesterone-mediated oocyte maturation 6.45 map04950 Maturity onset diabetes of the young 6.45 map05050 Dentatorubropallidoluysian atrophy (DRPLA) 6.45 map04140 Regulation of autophagy 6.45 map04940 Type I diabetes mellitus 6.45 map04910 Insulin signaling pathway 6.45 map04810 Regulation of actin cytoskeleton 6.45 map04930 Type II diabetes mellitus 4.30 map04510 Focal adhesion 4.30 map05214 Glioma 4.30 map04730 Long-term depression 4.30 map04115 p53 signaling pathway 4.30 map05218 Melanoma 1.08  map00380
map00380Tryptophan metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with IlGF domain which could be assigned to a KEGG orthologous group, and not all proteins containing IlGF domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of IlGF domains in PDB
PDB code Main view Title 1a7f 
INSULIN MUTANT B16 GLU, B24 GLY, DES-B30, NMR, 20 STRUCTURES 1ai0 
R6 HUMAN INSULIN HEXAMER (NON-SYMMETRIC), NMR, 10 STRUCTURES 1aiy 
R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 10 STRUCTURES 1aph 
CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 1b17 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 5.00 COORDINATES) 1b18 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 5.53 COORDINATES) 1b19 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 5.80 COORDINATES) 1b2a 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.00 COORDINATES) 1b2b 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.16 COORDINATES) 1b2c 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.26 COORDINATES) 1b2d 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.35 COORDINATES) 1b2e 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.50 COORDINATES) 1b2f 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 6.98 COORDINATES) 1b2g 
PH AFFECTS GLU B13 SWITCHING AND SULFATE BINDING IN CUBIC INSULIN CRYSTALS (PH 9.00 COORDINATES) 1b9e 
HUMAN INSULIN MUTANT SERB9GLU 1b9g 
INSULIN-LIKE-GROWTH-FACTOR-1 1ben 
INSULIN COMPLEXED WITH 4-HYDROXYBENZAMIDE 1bom 
THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN 1bon 
THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN 1bph 
CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 1bqt 
THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES 1bzv 
[D-ALAB26]-DES(B27-B30)-INSULIN-B26-AMIDE A SUPERPOTENT SINGLE-REPLACEMENT INSULIN ANALOGUE, NMR, MINIMIZED AVERAGE STRUCTURE 1cph 
CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 1dei 
DESHEPTAPEPTIDE (B24-B30) INSULIN 1dph 
CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 1efe 
AN ACTIVE MINI-PROINSULIN, M2PI 1ev3 
Structure of the rhombohedral form of the M-cresol/insulin R6 hexamer 1ev6 
Structure of the monoclinic form of the M-cresol/insulin R6 hexamer 1evr 
The structure of the resorcinol/insulin R6 hexamer 1fu2 
FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA 1fub 
FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA 1g7a 
1.2 A structure of T3R3 human insulin at 100 K 1g7b 
1.3 A STRUCTURE OF T3R3 HUMAN INSULIN AT 100 K 1guj 
Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. 1gzr 
Human Insulin-like growth factor; ESRF data 1gzy 
Human Insulin-like growth factor; In-house data 1gzz 
Human Insulin-like growth factor; Hamburg data 1h02 
Human Insulin-like growth factor; SRS Daresbury data 1h59 
Complex of IGFBP-5 with IGF-I 1hiq 
PARADOXICAL STRUCTURE AND FUNCTION IN A MUTANT HUMAN INSULIN ASSOCIATED WITH DIABETES MELLITUS 1his 
STRUCTURE AND DYNAMICS OF DES-PENTAPEPTIDE-INSULIN IN SOLUTION: THE MOLTEN-GLOBULE HYPOTHESIS 1hit 
RECEPTOR BINDING REDEFINED BY A STRUCTURAL SWITCH IN A MUTANT HUMAN INSULIN 1hls 
NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) 1ho0 
NEW B-CHAIN MUTANT OF BOVINE INSULIN 1htv 
CRYSTAL STRUCTURE OF DESTRIPEPTIDE (B28-B30) INSULIN 1hui 
INSULIN MUTANT (B1, B10, B16, B27)GLU, DES-B30, NMR, 25 STRUCTURES 1igl 
SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR II RELATIONSHIP TO RECEPTOR AND BINDING PROTEIN INTERACTIONS 1imx 
1.8 Angstrom crystal structure of IGF-1 1iog 
INSULIN MUTANT A3 GLY,(B1, B10, B16, B27)GLU, DES-B30, NMR, 19 STRUCTURES 1ioh 
INSULIN MUTANT A8 HIS,(B1, B10, B16, B27)GLU, DES-B30, NMR, 26 STRUCTURES 1iza 
ROLE OF B13 GLU IN INSULIN ASSEMBLY: THE HEXAMER STRUCTURE OF RECOMBINANT MUTANT (B13 GLU-> GLN) INSULIN 1izb 
ROLE OF B13 GLU IN INSULIN ASSEMBLY: THE HEXAMER STRUCTURE OF RECOMBINANT MUTANT (B13 GLU-> GLN) INSULIN 1j73 
Crystal structure of an unstable insulin analog with native activity. 1jca 
Non-standard Design of Unstable Insulin Analogues with Enhanced Activity 1jco 
Solution structure of the monomeric [Thr(B27)->Pro,Pro(B28)->Thr] insulin mutant (PT insulin) 1k3m 
NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES 1kmf 
NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES 1lkq 
NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES 1lph 
LYS(B28)PRO(B29)-HUMAN INSULIN 1m5a 
Crystal Structure of 2-Co(2+)-Insulin at 1.2A Resolution 1mhi 
THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN INSULIN DIMER. A STUDY OF THE B9(ASP) MUTANT OF HUMAN INSULIN USING NUCLEAR MAGNETIC RESONANCE DISTANCE GEOMETRY AND RESTRAINED MOLECULAR DYNAMICS 1mhj 
SOLUTION STRUCTURE OF THE SUPERACTIVE MONOMERIC DES-[PHE(B25)] HUMAN INSULIN MUTANT. ELUCIDATION OF THE STRUCTURAL BASIS FOR THE MONOMERIZATION OF THE DES-[PHE(B25)] INSULIN AND THE DIMERIZATION OF NATIVE INSULIN 1mpj 
X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL 1mso 
T6 Human Insulin at 1.0 A Resolution 1os3 
Dehydrated T6 human insulin at 100 K 1os4 
Dehydrated T6 human insulin at 295 K 1pid 
BOVINE DESPENTAPEPTIDE INSULIN 1pmx 
INSULIN-LIKE GROWTH FACTOR-I BOUND TO A PHAGE-DERIVED PEPTIDE 1q4v 
CRYSTAL STRUCTURE OF ALLO-ILEA2-INSULIN, AN INACTIVE CHIRAL ANALOGUE: IMPLICATIONS FOR THE MECHANISM OF RECEPTOR 1qiy 
HUMAN INSULIN HEXAMERS WITH CHAIN B HIS MUTATED TO TYR COMPLEXED WITH PHENOL 1qiz 
HUMAN INSULIN HEXAMERS WITH CHAIN B HIS MUTATED TO TYR COMPLEXED WITH RESORCINOL 1qj0 
HUMAN INSULIN HEXAMERS WITH CHAIN B HIS MUTATED TO TYR 1rwe 
Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues 1sdb 
PORCINE DESB1-2 DESPENTAPEPTIDE(B26-B30) INSULIN 1sf1 
NMR STRUCTURE OF HUMAN INSULIN under Amyloidogenic Condition, 15 STRUCTURES 1sjt 
MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES 1sju 
MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES 1t1k 
NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES 1t1p 
NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-THR, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES 1t1q 
NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ABA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES 1tgr 
Crystal Structure of mini-IGF-1(2) 1trz 
CRYSTALLOGRAPHIC EVIDENCE FOR DUAL COORDINATION AROUND ZINC IN THE T3R3 HUMAN INSULIN HEXAMER 1tyl 
THE STRUCTURE OF A COMPLEX OF HEXAMERIC INSULIN AND 4'-HYDROXYACETANILIDE 1tym 
THE STRUCTURE OF A COMPLEX OF HEXAMERIC INSULIN AND 4'-HYDROXYACETANILIDE 1uz9 
Crystallographic and solution studies of N-lithocholyl insulin: a new generation of prolonged-acting insulins. 1vkt 
HUMAN INSULIN TWO DISULFIDE MODEL, NMR, 10 STRUCTURES 1w8p 
Structural properties of the B25Tyr-NMe-B26Phe insulin mutant. 1wav 
CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN 1wqj 
Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) 1xda 
STRUCTURE OF INSULIN 1xgl 
HUMAN INSULIN DISULFIDE ISOMER, NMR, 10 STRUCTURES 1xw7 
Diabetes-Associated Mutations in Human Insulin: Crystal Structure and Photo-Cross-Linking Studies of A-Chain Variant Insulin Wakayama 1zeg 
STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL 1zeh 
STRUCTURE OF INSULIN 1zei 
CROSS-LINKED B28 ASP INSULIN 1zni 
INSULIN 1znj 
INSULIN, MONOCLINIC CRYSTAL FORM 2a3g 
The structure of T6 bovine insulin 2aiy 
R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES 2bn1 
Insulin after a high dose x-ray burn 2bn3 
Insulin before a high dose x-ray burn 2c8q 
insuline(1sec) and UV laser excited fluorescence 2c8r 
insuline(60sec) and UV laser excited fluorescence 2ceu 
Despentapeptide insulin in acetic acid (pH 2) 2dsp 
Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins 2dsq 
Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins 2dsr 
Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins 2efa 
Neutron crystal structure of cubic insulin at pD6.6 2g4m 
Insulin collected at 2.0 A wavelength 2g54 
Crystal structure of Zn-bound human insulin-degrading enzyme in complex with insulin B chain 2g56 
crystal structure of human insulin-degrading enzyme in complex with insulin B chain 2gf1 
SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR 1: A NUCLEAR MAGNETIC RESONANCE AND RESTRAINED MOLECULAR DYNAMICS STUDY 2h67 
NMR structure of human insulin mutant HIS-B5-ALA, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures 2hh4 
NMR structure of human insulin mutant GLY-B8-D-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures 2hho 
NMR structure of human insulin mutant GLY-B8-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures 2hiu 
NMR STRUCTURE OF HUMAN INSULIN IN 20% ACETIC ACID, ZINC-FREE, 10 STRUCTURES 2ins 
THE STRUCTURE OF DES-PHE B1 BOVINE INSULIN 2jmn 
NMR structure of human insulin mutant His-B10-Asp, Pro-B28-Lys, Lys-B29-Pro, 20 structures 2jum 
ThrA3-DKP-insulin 2juu 
allo-ThrA3 DKP-insulin 2juv 
AbaA3-DKP-insulin 2jv1 
NMR structure of human insulin monomer in 35% CD3CN zinc free, 50 structures 2jzq 
Design of an Active Ultra-Stable Single-Chain Insulin Analog 20 Structures 2k91 
Enhancing the activity of insulin by stereospecific unfolding 2k9r 
Enhancing the activity of insulin by stereospecific unfolding 2kji 
A divergent ins protein in c. elegans structurally resemble insulin and activates the human insulin receptor 2kjj 
Dynamics of insulin probed by 1H-NMR amide proton exchange anomalous flexibility of the receptor-binding surface 2kju 
NMR structure of human insulin mutant glu-b21-d-glu, his-b10 asp pro-b28-lys, lys-b29-pro, 20 structures 2kqp 
NMR Structure of Proinsulin 2kqq 
NMR structure of human insulin mutant gly-b8-d-ala, his-b10-asp, pro-b28-lys, lys-b29-pro, 20 structures 2kxk 
Human Insulin Mutant A22Gly-B31Lys-B32Arg 2l1y 
NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures 2l1z 
NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures 2l29 
Complex structure of E4 mutant human IGF2R domain 11 bound to IGF-II 2lgb 
Modified A22Gly-B31Arg Human Insulin 2lwz 
NMR Structures of Single-chain Insulin 2m1d 
Biosynthetic engineered B28K-B29P human insulin monomer structure in in water/acetonitrile solutions. 2m1e 
Biosynthetic engineered B28K-B29P human insulin monomer structure in in water solutions. 2m2m 
Structure of [L-HisB24] insulin analogue at pH 1.9 2m2n 
Structure of [L-HisB24] insulin analogue at pH 8.0 2m2o 
Structure of [D-HisB24] insulin analogue at pH 1.9 2m2p 
Structure of [D-HisB24] insulin analogue at pH 8.0 2mli 
2MLI 2mpg 
2MPG 2mpi 
2MPI 2mvc 
2MVC 2mvd 
2MVD 2n2v 
2N2V 2n2w 
2N2W 2n2x 
2N2X 2oly 
Structure of human insulin in presence of urea at pH 7.0 2olz 
Structure of human insulin in presence of thiocyanate at pH 7.0 2om0 
Structure of human insulin in presence of urea at pH 6.5 2om1 
Structure of human insulin in presence of thiocyanate at pH 6.5 2omg 
Structure of human insulin cocrystallized with protamine and urea 2omh 
Structure of human insulin cocrystallized with ARG-12 peptide in presence of urea 2omi 
Structure of human insulin cocrystallized with protamine 2qiu 
Structure of Human Arg-Insulin 2r34 
Crystal structure of MN human arg-insulin 2r35 
Crystal structure of RB human arg-insulin 2r36 
Crystal structure of ni human ARG-insulin 2rn5 
Humal Insulin Mutant B31Lys-B32Arg 2tci 
X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL 2v5p 
COMPLEX STRUCTURE OF HUMAN IGF2R DOMAINS 11-13 BOUND TO IGF-II 2vjz 
Crystal structure form ultalente insulin microcrystals 2vk0 
Crystal structure form ultalente insulin microcrystals 2w44 
Structure DeltaA1-A4 insulin 2wby 
Crystal structure of human insulin-degrading enzyme in complex with insulin 2wc0 
crystal structure of human insulin degrading enzyme in complex with iodinated insulin 2wru 
Semi-synthetic highly active analogue of human insulin NMeAlaB26-DTI- NH2 2wrv 
Semi-synthetic highly active analogue of human insulin NMeHisB26-DTI- NH2 2wrw 
Semi-synthetic highly active analogue of human insulin D-ProB26-DTI- NH2 2wrx 
Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 3.0 2ws0 
Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 7.5 2ws1 
Semi-synthetic analogue of human insulin NMeTyrB26-insulin in monomer form 2ws4 
Semi-synthetic analogue of human insulin ProB26-DTI in monomer form 2ws6 
Semi-synthetic analogue of human insulin NMeTyrB26-insulin in hexamer form 2ws7 
Semi-synthetic analogue of human insulin ProB26-DTI 2zp6 
Crystal structure of Bovine Insulin (Hexameric form) 2zpp 
Neutron crystal structure of cubic insulin at pD9 3aiy 
R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE 3bxq 
The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition 3e4z 
Crystal structure of human insulin degrading enzyme in complex with insulin-like growth factor II 3e7y 
Structure of human insulin 3e7z 
Structure of human insulin 3exx 
Structure of the T6 human insulin derivative with nickel at 1.35 A resolution 3fhp 
A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution 3fq9 
Design of an insulin analog with enhanced receptor-binding selectivity. Rationale, structure, and therapeutic implications 3gf1 
SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR 1: A NUCLEAR MAGNETIC RESONANCE AND RESTRAINED MOLECULAR DYNAMICS STUDY 3gky 
The Structural Basis of an ER Stress-Associated Bottleneck in a Protein Folding Landscape 3i3z 
Human insulin 3i40 
Human insulin 3ilg 
Crystal structure of humnan insulin Sr+2 complex 3inc 
Crystal structure of human insulin with Ni+2 complex 3ins 
STRUCTURE OF INSULIN. RESULTS OF JOINT NEUTRON AND X-RAY REFINEMENT 3ir0 
Crystal Structure of Human Insulin complexed with Cu+2 metal ion 3jsd 
Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus 3kq6 
Enhancing the Therapeutic Properties of a Protein by a Designed Zinc-Binding Site, Structural principles of a novel long-acting insulin analog 3kr3 
Crystal structure of IGF-II antibody complex 3lri 
Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I 3mth 
X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL 3p2x 
Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity 3p33 
Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity 3q6e 
Human insulin in complex with cucurbit[7]uril 3rov 
Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus 3rto 
Acoustically mounted porcine insulin microcrystals yield an X-ray SAD structure 3t2a 
TMAO-grown cubic insulin (porcine) 3tt8 
Crystal Structure Analysis of Cu Human Insulin Derivative 3u4n 
A novel covalently linked insulin dimer 3v19 
Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health 3v1g 
Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health 3w11 
Insulin receptor ectodomain construct comprising domains L1-CR in complex with human insulin, Alpha-CT peptide(704-719) and FAB 83-7 3w12 
Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alpha-CT peptide(704-719) and FAB 83-7 3w13 
Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alphact peptide(693-719) and FAB 83-7 3w14 
Insulin receptor ectodomain construct comprising domains L1,CR,L2, FNIII-1 and alphact peptide in complex with bovine insulin and FAB 83-14 3w7y 
0.92A structure of 2Zn human insulin at 100K 3w7z 
1.15A structure of human 2Zn insulin at 293K 3w80 
Crystal structure of dodecamer human insulin with double C-axis length of the hexamer 2 Zn insulin cell 3zi3 
Crystal structure of the B24His-insulin - human analogue 3zqr 
NMePheB25 insulin analogue crystal structure 3zs2 
TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure 3zu1 
Structure of LysB29(Nepsilon omega-carboxyheptadecanoyl) des(B30) Human Insulin 4a7e 
X-ray crystal structure of porcine insulin flash-cooled at high pressure 4aiy 
R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'GREEN' SUBSTATE, AVERAGE STRUCTURE 4ajx 
Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin 4ajz 
Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin 4ak0 
Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin 4akj 
Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin 4bs3 
Bovin insulin structure determined by in situ crystal analysis and sulfur-SAD phasing at room temperature 4cxl 
4CXL 4cxn 
4CXN 4cy7 
4CY7 4e7t 
The structure of T6 bovine insulin 4e7u 
The structure of T3R3 bovine insulin 4e7v 
The structure of R6 bovine insulin 4efx 
Highly biologically active insulin with additional disulfide bond 4eww 
Human Insulin 4ewx 
Human Insulin 4ewz 
Human Insulin 4ex0 
Human Insulin 4ex1 
Human Insulin 4exx 
Human Insulin 4ey1 
Human Insulin 4ey9 
Human Insulin 4eyd 
Human Insulin 4eyn 
Human Insulin 4eyp 
Human Insulin 4f0n 
Human Insulin 4f0o 
Human Insulin 4f1a 
Human Insulin 4f1b 
Human Insulin 4f1c 
Human Insulin 4f1d 
Human Insulin 4f1f 
Human Insulin 4f1g 
Human insulin 4f4t 
Human Insulin 4f4v 
Human Insulin 4f51 
Human Insulin 4f8f 
Human Insulin 4fg3 
Crystal Structure Analysis of the Human Insulin 4fka 
High resolution structure of the manganese derivative of insulin 4gbc 
Crystal structure of aspart insulin at pH 6.5 4gbi 
Crystal structure of aspart insulin at pH 6.5 4gbk 
Crystal structure of aspart insulin at pH 8.5 4gbl 
Crystal structure of aspart insulin at pH 8.5 4gbn 
Crystal structure of aspart insulin at pH 6.5 4i5y 
Insulin protein crystallization via langmuir-blodgett 4i5z 
Insulin protein crystallization via langmuir-blodgett 4idw 
Polycrystalline T6 Bovine Insulin: Anisotropic Lattice Evolution and Novel Structure Refinement Strategy 4ihn 
High Resolution Insulin by Langmuir-Blodgett Modified Hanging Drop Vapour Diffusion 4ins 
THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION 4iuz 
High resolution crystal structure of racemic ester insulin 4iyd 
Insulin glargine crystal structure 1 4iyf 
Insulin glargine crystal structure 2 4m4f 
Radiation damage study of Cu T6-insulin - 0.01 MGy 4m4h 
Radiation damage study of Cu T6-insulin - 0.06 MGy 4m4i 
Radiation damage study of Cu T6-insulin - 0.12 MGy 4m4j 
Radiation damage study of Cu T6-insulin - 0.30 MGy 4m4l 
The structure of Cu T6 bovine insulin 4m4m 
The structure of Ni T6 bovine insulin 4nib 
4NIB 4oga 
4OGA 4p65 
4P65 4q5z 
4Q5Z 4rxw 
4RXW 4une 
4UNE 4ung 
4UNG 4unh 
4UNH 4xc4 
4XC4 4xss 
4XSS 5aiy 
R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'RED' SUBSTATE, AVERAGE STRUCTURE 5boq 
5BOQ 5bpo 
5BPO 5bqq 
5BQQ 5bts 
5BTS 5cjo 
5CJO 5cny 
5CNY 5co2 
5CO2 5co6 
5CO6 5co9 
5CO9 5d52 
5D52 5d53 
5D53 5d54 
5D54 5d5e 
5D5E 5e7w 
5E7W 5ems 
5EMS 5en9 
5EN9 5ena 
5ENA 5fb6 
5FB6 5l3l 
5L3L 5l3m 
5L3M 5l3n 
5L3N 5lis 
5LIS 6ins 
X-RAY ANALYSIS OF THE SINGLE CHAIN B29-A1 PEPTIDE-LINKED INSULIN MOLECULE. A COMPLETELY INACTIVE ANALOGUE 7ins 
STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z 9ins 
MONOVALENT CATION BINDING IN CUBIC INSULIN CRYSTALS - Links (links to other resources describing this domain)
-
PROSITE IlGF_DOMAIN PFAM ins INTERPRO IPR016179

