PBPbBacterial periplasmic substrate-binding proteins |
|---|
| SMART accession number: | SM00062 |
|---|---|
| Description: | bacterial proteins, eukaryotic ones are in PBPe |
| Interpro abstract (IPR001638): | Bacterial high affinity transport systems are involved in active transport of solutes across the cytoplasmic membrane. Most of the bacterial ABC (ATP-binding cassette) importers are composed of one or two transmembrane permease proteins, one or two nucleotide-binding proteins and a highly specific periplasmic solute-binding protein. In Gram-negative bacteria the solute-binding proteins are dissolved in the periplasm, while in archaea and Gram-positive bacteria, their solute-binding proteins are membrane-anchored lipoproteins [ (PUBMED:8003968) (PUBMED:18310026) ]. On the basis of sequence similarities, the vast majority of these solute-binding proteins can be grouped [ (PUBMED:8336670) ] into eight families or clusters, which generally correlate with the nature of the solute bound. This entry represents a domain found in the solute-binding protein family 3 members from Gram-positive bacteria, Gram-negative bacteria and archaea. This domain can also be found in the N-terminal of the membrane-bound lytic murein transglycosylase F (MltF) protein. MltF is a murein-degrading enzyme that degrades murein glycan strands and insoluble, high-molecular weight murein sacculi, with the concomitant formation of a 1,6-anhydromuramoyl product [ (PUBMED:18234673) ]. Familiy 3 members include:
|
| Family alignment: |
There are 124608 PBPb domains in 119061 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing PBPb domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with PBPb domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing PBPb domain in the selected taxonomic class.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
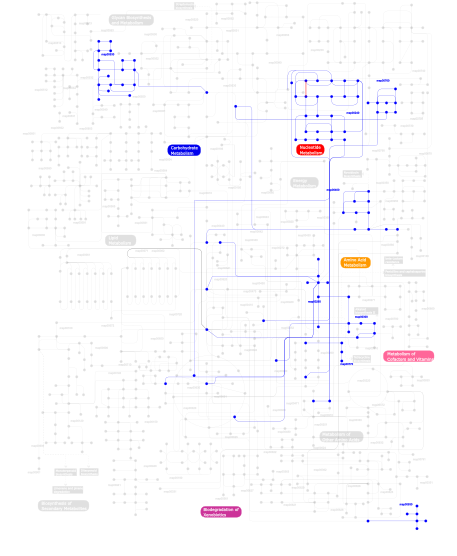
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 85.59 map02010 ABC transporters - General 6.15 map02020 Two-component system - General 3.87 map02040 Flagellar assembly 3.16  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 0.18  map00260
map00260Glycine, serine and threonine metabolism 0.18  map00770
map00770Pantothenate and CoA biosynthesis 0.18  map00930
map00930Caprolactam degradation 0.18  map00760
map00760Nicotinate and nicotinamide metabolism 0.18  map00300
map00300Lysine biosynthesis 0.18  map00240
map00240Pyrimidine metabolism 0.18  map00530
map00530Aminosugars metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with PBPb domain which could be assigned to a KEGG orthologous group, and not all proteins containing PBPb domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of PBPb domains in PDB
PDB code Main view Title 1ftj 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH GLUTAMATE AT 1.9 RESOLUTION 1ftk 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2I) IN COMPLEX WITH KAINATE AT 1.6 A RESOLUTION 1ftl 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH THE ANTAGONIST DNQX AT 1.8 A RESOLUTION 1ftm 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH AMPA AT 1.7 RESOLUTION 1fto 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN THE APO STATE AT 2.0 A RESOLUTION 1fw0 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH KAINATE AT 2.0 A RESOLUTION 1ggg 
GLUTAMINE BINDING PROTEIN OPEN LIGAND-FREE STRUCTURE 1gr2 
STRUCTURE OF A GLUTAMATE RECEPTOR LIGAND BINDING CORE (GLUR2) COMPLEXED WITH KAINATE 1hpb 
THE BACTERIAL PERIPLASMIC HISTIDINE-BINDING PROTEIN: STRUCTURE(SLASH)FUNCTION ANALYSIS OF THE LIGAND-BINDING SITE AND COMPARISON WITH RELATED PROTEINS 1hsl 
REFINED 1.89 ANGSTROMS STRUCTURE OF THE HISTIDINE-BINDING PROTEIN COMPLEXED WITH HISTIDINE AND ITS RELATIONSHIP WITH MANY OTHER ACTIVE TRANSPORT(SLASH)CHEMOSENSORY RECEPTORS 1ii5 
CRYSTAL STRUCTURE OF THE GLUR0 LIGAND BINDING CORE COMPLEX WITH L-GLUTAMATE 1iit 
GLUR0 LIGAND BINDING CORE COMPLEX WITH L-SERINE 1iiw 
GLUR0 LIGAND BINDING CORE: CLOSED-CLEFT LIGAND-FREE STRUCTURE 1laf 
STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN 1lag 
STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN 1lah 
STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN 1lb8 
Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution 1lb9 
Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution 1lbb 
Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution 1lbc 
Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution 1lst 
THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND 1m5b 
X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. 1m5c 
X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION 1m5d 
X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION 1m5e 
X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION 1m5f 
X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION 1mm6 
crystal structure of the GluR2 ligand binding core (S1S2J) in complex with quisqualate in a non zinc crystal form at 2.15 angstroms resolution 1mm7 
Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Quisqualate in a Zinc Crystal Form at 1.65 Angstroms Resolution 1mqd 
X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.46 A resolution. Crystallization in the presence of lithium sulfate. 1mqg 
Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution 1mqh 
Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution 1mqi 
Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution 1mqj 
Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution 1ms7 
X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate 1mxu 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) in complex with bromo-willardiine (Control for the crystal titration experiments) 1mxv 
crystal titration experiments (AMPA co-crystals soaked in 10 mM BrW) 1mxw 
crystal titration experiments (AMPA co-crystals soaked in 1 mM BrW) 1mxx 
crystal titration experiments (AMPA co-crystals soaked in 100 uM BrW) 1mxy 
crystal titration experiments (AMPA co-crystals soaked in 10 uM BrW) 1mxz 
crystal titration experiments (AMPA co-crystals soaked in 1 uM BrW) 1my0 
crystal titration experiments (AMPA co-crystals soaked in 100 nM BrW) 1my1 
crystal titration experiments (AMPA co-crystals soaked in 10 nM BrW) 1my2 
crystal titration experiment (AMPA complex control) 1my3 
crystal structure of glutamate receptor ligand-binding core in complex with bromo-willardiine in the Zn crystal form 1my4 
crystal structure of glutamate receptor ligand-binding core in complex with iodo-willardiine in the Zn crystal form 1n0t 
X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. 1nnk 
X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. 1nnp 
X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. 1p1n 
GluR2 Ligand Binding Core (S1S2J) Mutant L650T in Complex with Kainate 1p1o 
Crystal structure of the GluR2 ligand-binding core (S1S2J) mutant L650T in complex with quisqualate 1p1q 
Crystal structure of the GluR2 ligand binding core (S1S2J) L650T mutant in complex with AMPA 1p1u 
Crystal structure of the GluR2 ligand-binding core (S1S2J) L650T mutant in complex with AMPA (ammonium sulfate crystal form) 1p1w 
Crystal structure of the GluR2 ligand-binding core (S1S2J) with the L483Y and L650T mutations and in complex with AMPA 1pb7 
CRYSTAL STRUCTURE OF THE NR1 LIGAND BINDING CORE IN COMPLEX WITH GLYCINE AT 1.35 ANGSTROMS RESOLUTION 1pb8 
CRYSTAL STRUCTURE OF THE NR1 LIGAND BINDING CORE IN COMPLEX WITH D-SERINE AT 1.45 ANGSTROMS RESOLUTION 1pb9 
CRYSTAL STRUCTURE OF THE NR1 LIGAND BINDING CORE IN COMPLEX WITH D-CYCLOSERINE AT 1.60 ANGSTROMS RESOLUTION 1pbq 
CRYSTAL STRUCTURE OF THE NR1 LIGAND BINDING CORE IN COMPLEX WITH 5,7-DICHLOROKYNURENIC ACID (DCKA) AT 1.90 ANGSTROMS RESOLUTION 1s50 
X-ray structure of the GluR6 ligand binding core (S1S2A) in complex with glutamate at 1.65 A resolution 1s7y 
Crystal structure of the GluR6 ligand binding core in complex with glutamate at 1.75 A resolution orthorhombic form 1s9t 
Crystal structure of the GLUR6 ligand binding core in complex with quisqualate at 1.8A resolution 1sd3 
Crystal structure of the GLUR6 ligand binding core in complex with 2S,4R-4-methylglutamate at 1.8 Angstrom resolution 1syh 
X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. 1syi 
X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. 1tt1 
CRYSTAL STRUCTURE OF THE GLUR6 LIGAND BINDING CORE IN COMPLEX WITH KAINATE 1.93 A RESOLUTION 1txf 
CRYSTAL STRUCTURE OF THE GLUR5 LIGAND BINDING CORE IN COMPLEX WITH GLUTAMATE AT 2.1 ANGSTROM RESOLUTION 1vso 
Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution 1wdn 
GLUTAMINE-BINDING PROTEIN 1wvj 
Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP 1xhy 
X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution 1xt8 
Crystal Structure of Cysteine-Binding Protein from Campylobacter jejuni at 2.0 A Resolution 1y1m 
Crystal structure of the NR1 ligand binding core in complex with cycloleucine 1y1z 
Crystal structure of the NR1 ligand binding core in complex with ACBC 1y20 
Crystal structure of the NR1 ligand-binding core in complex with ACPC 1ycj 
Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate 2a5s 
Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate 2a5t 
Crystal Structure Of The NR1/NR2A ligand-binding cores complex 2aix 
X-ray structure of the GLUR2 ligand-binding core (S1S2J) in complex with (s)-thio-atpa at 2.2 a resolution. 2al4 
CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. 2al5 
Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with fluoro-willardiine and aniracetam 2anj 
Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution 2cmo 
The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209. 2f34 
Crystal Structure of the GluR5 Ligand Binding Core Dimer with UBP310 At 1.74 Angstroms Resolution 2f35 
Crystal Structure of the GluR5 Ligand Binding Core with UBP302 At 1.87 Angstroms Resolution 2f36 
Crystal Structure of the GluR5 Ligand Binding Core Dimer with Glutamate At 2.1 Angstroms Resolution 2gfe 
Crystal structure of the GluR2 A476E S673D Ligand Binding Core Mutant at 1.54 Angstroms Resolution 2i0b 
Crystal structure of the GluR6 ligand binding core ELKQ mutant dimer at 1.96 Angstroms Resolution 2i0c 
Crystal structure of the GluR6 ligand binding core dimer crosslinked by disulfide bonds between Y490C and L752C at 2.25 Angstroms Resolution 2i3v 
Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant 2i3w 
Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant 2ia4 
Crystal structure of Novel amino acid binding protein from Shigella flexneri 2iee 
Crystal Structure of YCKB_BACSU from Bacillus subtilis. Northeast Structural Genomics Consortium target SR574. 2lao 
THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND 2m8c 
The solution NMR structure of E. coli apo-HisJ 2o1m 
Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572 2ojt 
Structure and mechanism of kainate receptor modulation by anions 2p2a 
X-ray structure of the GluR2 ligand binding core (S1S2J) in complex with 2-Bn-tet-AMPA at 2.26A resolution 2pbw 
Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution 2pvu 
Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus 2pyy 
Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate 2q2a 
Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus 2q2c 
Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus 2q88 
Crystal structure of EhuB in complex with ectoine 2q89 
Crystal structure of EhuB in complex with hydroxyectoine 2qs1 
Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution 2qs2 
Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution 2qs3 
Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution 2qs4 
Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution 2rc7 
Crystal structure of the NR3A ligand binding core complex with glycine at 1.58 Angstrom resolution 2rc8 
Crystal structure of the NR3A ligand binding core complex with D-serine at 1.45 Angstrom resolution 2rc9 
Crystal structure of the NR3A ligand binding core complex with ACPC at 1.96 Angstrom resolution 2rca 
Crystal structure of the NR3B ligand binding core complex with glycine at 1.58 Angstrom resolution 2rcb 
Crystal structure of the NR3B ligand binding core complex with D-serine at 1.62 Angstrom resolution 2uxa 
Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. 2v25 
Structure of the Campylobacter jejuni antigen Peb1A, an aspartate and glutamate receptor with bound aspartate 2v3t 
Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form 2v3u 
Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine 2vha 
DEBP 2wky 
Crystal structure of the ligand-binding core of GluR5 in complex with the agonist 4-AHCP 2x26 
Crystal structure of the periplasmic aliphatic sulphonate binding protein SsuA from Escherichia coli 2xhd 
Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor 2xx7 
Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-( trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. 2xx8 
Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. 2xx9 
Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. 2xxh 
Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-( trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. 2xxi 
Crystal structure of 1-((4-(3-(trifluoromethyl)-6,7-dihydropyrano(4,3- c(pyrazol-1(4H)-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.6A resolution. 2xxr 
Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with glutamate 2xxt 
Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with kainate 2xxu 
Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with glutamate 2xxv 
Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with kainate 2xxw 
Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate 2xxx 
Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate (P21 21 21) 2xxy 
Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with kainate 2y7i 
Structural basis for high arginine specificity in Salmonella typhimurium periplasmic binding protein STM4351. 2yjp 
Crystal structure of the solute receptors for L-cysteine of Neisseria gonorrhoeae 2yln 
Crystal structure of the L-cystine solute receptor of Neisseria gonorrhoeae in the closed conformation 2zns 
Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with glutamate 2znt 
Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, dysiherbaine 2znu 
Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, neodysiherbaine A 3b6q 
Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) Mutant T686A in Complex with Glutamate at 2.0 Resolution 3b6t 
Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution 3b6w 
Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686S Mutant in Complex with Glutamate at 1.7 Resolution 3b7d 
Crystal structure of the GLUR2 ligand binding core (HS1S2J) in complex with CNQX at 2.5 A resolution 3bbr 
Crystal structure of the iGluR2 ligand binding core (S1S2J-N775S) in complex with a dimeric positive modulator as well as glutamate at 2.25 A resolution 3bft 
Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution 3bfu 
Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution 3bki 
Crystal Structure of the GluR2 ligand binding core (S1S2J) in complex with FQX at 1.87 Angstroms 3c31 
Crystal structure of GluR5 ligand-binding core in complex with lithium at 1.49 Angstrom resolution 3c32 
Crystal structure of GluR5 ligand-binding core in complex with sodium at 1.72 Angstrom resolution 3c33 
Crystal structure of GluR5 ligand-binding core in complex with potassium at 1.78 Angstrom resolution 3c34 
Crystal structure of GluR5 ligand-binding core in complex with rubidium at 1.82 Angstrom resolution 3c35 
Crystal structure of GluR5 ligand-binding core in complex with cesium at 1.97 Angstrom resolution 3c36 
Crystal structure of GluR5 ligand-binding core in complex with ammonium ions at 1.68 Angstrom resolution 3del 
The structure of CT381, the arginine binding protein from the periplasm Chlamydia trachomatis 3dln 
Crystal structure of the binding domain of the AMPA subunit GluR3 bound to glutamate 3dp4 
Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA 3dp6 
Crystal structure of the binding domain of the AMPA subunit GluR2 bound to glutamate 3e4r 
Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES 3en3 
Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate 3epe 
Crystal Structure of the GluR4 Ligand-Binding domain in complex with glutamate 3fas 
X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-glutamate at 1.40A resolution 3fat 
X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-AMPA at 1.90A resolution 3fuz 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with L-glutamate in space group P1 3fv1 
Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 3fv2 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with neodysiherbaine A in space group P1 3fvg 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 3fvk 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-deoxy-neodysiherbaine A in space group P1 3fvn 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 9-deoxy-neodysiherbaine A in space group P1 3fvo 
Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-epi-neodysiherbaine A in space group P1 3g3f 
Crystal structure of the GluR6 ligand binding domain dimer with glutamate and NaCl at 1.38 Angstrom resolution 3g3g 
Crystal structure of the GluR6 ligand binding domain dimer K665R mutant with glutamate and NaCl at 1.3 Angstrom resolution 3g3h 
Crystal structure of the GluR6 ligand binding domain dimer K665R I749L Q753K mutant with glutamate and NaCl at 1.5 Angstrom resolution 3g3i 
Crystal structure of the GluR6 ligand binding domain dimer I442H K494E I749L Q753K mutant with glutamate and NaCl at 1.37 Angstrom resolution 3g3j 
Crystal structure of the GluR6 ligand binding domain dimer I442H K494E K665R I749L Q753K mutant with glutamate and NaCl at 1.32 Angstrom resolution 3g3k 
Crystal structure of the GluR6 ligand binding domain dimer I442H K494E K665R I749L Q753K E757Q mutant with glutamate and NaCl at 1.24 Angstrom resolution 3gba 
X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution 3gbb 
X-ray strucutre of iGluR5 ligand-binding core (S1S2) in complex with MSVIII-19 at 2.10A resolution 3h03 
Crystal structure of the binding domain of the AMPA subunit GluR2 bound to UBP277 3h06 
Crystal structure of the binding domain of the AMPA subunit GluR2 bound to the willardiine antagonist, UBP282 3h6t 
Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution 3h6u 
Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution 3h6v 
Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution 3h6w 
Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution 3h7m 
Crystal Structure of a Histidine Kinase Sensor Domain with Similarity to Periplasmic Binding Proteins 3hv1 
Crystal structure of a polar amino acid ABC uptake transporter substrate binding protein from Streptococcus thermophilus 3i6v 
Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine 3ijo 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide 3ijx 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide 3ik6 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, chlorothiazide 3il1 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 3ilt 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide 3ilu 
Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydroflumethiazide 3k4u 
CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine 3kbr 
The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01 3kei 
Crystal Structure of the GluA4 Ligand-Binding domain L651V mutant in complex with glutamate 3kfm 
Crystal Structure of the GluA4 Ligand-Binding domain L651V mutant in complex with kainate 3kgc 
Isolated ligand binding domain dimer of GluA2 ionotropic glutamate receptor in complex with glutamate, LY 404187 and ZK 200775 3ksj 
The alkanesulfonate-binding protein SsuA from Xabthomonas axonopodis pv. citri bound to MES 3ksx 
The alkanesulfonate-binding protein SsuA from Xanthomonas axonopodis pv. citri bound to MOPS 3kzg 
Crystal structure of an arginine 3rd transport system periplasmic binding protein from Legionella pneumophila 3lsf 
Piracetam bound to the ligand binding domain of GluA2 3lsl 
Piracetam bound to the ligand binding domain of GluA2 (flop form) 3lsw 
Aniracetam bound to the ligand binding domain of GluA3 3lsx 
Piracetam bound to the ligand binding domain of GluA3 3m3f 
PEPA bound to the ligand binding domain of GluA3 (flop form) 3m3k 
Ligand binding domain (S1S2) of GluA3 (flop) 3m3l 
PEPA bound to the ligand binding domain of GluA2 (flop form) 3mpk 
Crystal Structure of Bordetella pertussis BvgS periplasmic VFT2 domain 3mpl 
Crystal Structure of Bordetella pertussis BvgS VFT2 domain (Double Mutant F375E/Q461E) 3n26 
Cpn0482 : the arginine binding protein from the periplasm of chlamydia Pneumoniae 3o28 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3o29 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3o2a 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3o6g 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3o6h 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3o6i 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3oek 
Crystal structure of GluN2D ligand-binding core in complex with L-aspartate 3oel 
Crystal structure of GluN2D ligand-binding core in complex with D-glutamate 3oem 
Crystal structure of GluN2D ligand-binding core in complex with N-methyl-D-aspartate 3oen 
Crystal structure of GluN2D ligand-binding core in complex with L-glutamate 3pd8 
X-ray structure of the ligand-binding core of GluA2 in complex with (S)-7-HPCA at 2.5 A resolution 3pd9 
X-ray structure of the ligand-binding core of GluA2 in complex with (R)-5-HPCA at 2.1 A resolution 3pmv 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3pmw 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3pmx 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 3qax 
Crystal structure anlysis of the cpb0502 3qsl 
Structure of CAE31940 from Bordetella bronchiseptica RB50 3qxm 
Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A 3r7x 
Crystal Structure Analysis of a Quinazolinedione sulfonamide bound to human GluR2: A Novel Class of Competitive AMPA Receptor Antagonists with Oral Activity 3rn8 
Crystal Structure of iGluR2 Ligand Binding Domain and Symmetrical Carboxyl Containing Potentiator 3rnn 
Crystal Structure of iGluR2 Ligand Binding Domain with Symmetric Sulfonamide Containing Potentiator 3rt6 
Fluorowillardiine bound to the ligand binding domain of GluA3 3rt8 
Chlorowillardiine bound to the ligand binding domain of GluA3 3rtf 
Chlorowillardiine bound to the ligand binding domain of GluA2 3rtw 
Nitrowillardiine bound to the ligand binding domain of GluA2 3s2v 
Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution 3s9e 
Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate 3t93 
Glutamate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 3t96 
Iodowillardiine bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 3t9h 
Kainate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 3t9u 
CNQX bound to an oxidized double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 3t9v 
CNQX bound to a reduced double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 3t9x 
Glutamate bound to a double cysteine mutant (V484C/E657C) of the ligand binding domain of GluA2 3tdj 
Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-97 at 1.95 A resolution 3tkd 
Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and cyclothiazide at 1.45 A resolution 3tql 
Structure of the amino acid ABC transporter, periplasmic amino acid-binding protein from Coxiella burnetii. 3tza 
Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 1.9A resolution 3u92 
Crystal structure of the GluK3 ligand binding domain complex with kainate and zinc: P2221 form 3u93 
Crystal structure of the GluK3 ligand binding domain complex with glutamate and zinc: P2221 form 3u94 
Crystal structure of the GluK3 ligand binding domain complex with glutamate and zinc: P21212 form 3ua8 
Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 3vv5 
Crystal structure of TTC0807 complexed with (S)-2-aminoethyl-L-cysteine (AEC) 3vvd 
Crystal structure of TTC0807 complexed with Ornithine 3vve 
Crystal structure of TTC0807 complexed with Lysine 3vvf 
Crystal structure of TTC0807 complexed with Arginine 3zsf 
Crystal structure of the L-cystine solute receptor of Neisseria gonorrhoeae in the unliganded open conformation 4bdl 
Crystal structure of the GluK2 K531A LBD dimer in complex with glutamate 4bdm 
Crystal structure of the GluK2 K531A LBD dimer in complex with kainate 4bdn 
Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with glutamate 4bdo 
Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with kainate 4bdq 
Crystal structure of the GluK2 R775A LBD dimer in complex with glutamate 4bdr 
Crystal structure of the GluK2 R775A LBD dimer in complex with kainate 4c0r 
Molecular and structural basis of glutathione import in Gram-positive bacteria via GshT and the cystine ABC importer TcyBC of Streptococcus mutans. 4dld 
Crystal structure of the GluK1 ligand-binding domain (S1S2) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 2.0 A resolution 4dz1 
Crystal structure of DalS, an ATP binding cassette transporter for D-alanine from Salmonella enterica 4e0w 
Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate 4e0x 
Crystal structure of the kainate receptor GluK1 ligand-binding domain in complex with kainate in the absence of glycerol 4eq9 
1.4 Angstrom Crystal Structure of ABC Transporter Glutathione-Binding Protein GshT from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Glutathione 4f1y 
CNQX bound to the ligand binding domain of GluA3 4f22 
Kainate bound to the K660A mutant of the ligand binding domain of GluA3 4f29 
Quisqualate bound to the ligand binding domain of GluA3i 4f2o 
Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 4f2q 
Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 4f31 
Kainate bound to the D655A mutant of the ligand binding domain of GluA3 4f39 
Kainate bound to the ligand binding domain of GluA3 4f3b 
Glutamate bound to the D655A mutant of the ligand binding domain of GluA3 4f3g 
Kainate bound to the ligand binding domain of GluA3i 4f3p 
Crystal structure of a Glutamine-binding periplasmic protein from Burkholderia pseudomallei in complex with glutamine 4f3s 
Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica 4fat 
Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator 4g4p 
Crystal structure of glutamine-binding protein from Enterococcus faecalis at 1.5 A 4g8m 
Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist CBG-IV at 2.05A resolution 4g8n 
Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M 4gvo 
Putative L-Cystine ABC transporter from Listeria monocytogenes 4gxs 
Ligand binding domain of GluA2 (AMPA/glutamate receptor) bound to (-)-kaitocephalin 4h5f 
Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1 4h5g 
Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 2 4h8i 
Structure of GluK2-LBD in complex with GluAzo 4h8j 
Structure of GluA2-LBD in complex with MES 4i62 
1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine 4igr 
Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist ZA302 4igt 
Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution 4io2 
Crystal Structure of the AvGluR1 ligand binding domain complex with glutamate at 1.37 Angstrom resolution 4io3 
Crystal Structure of the AvGluR1 ligand binding domain complex with aspartate at 1.66 Angstrom resolution 4io4 
Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution 4io5 
Crystal Structure of the AvGluR1 ligand binding domain complex with alanine at 1.72 Angstrom resolution 4io6 
Crystal Structure of the AvGluR1 ligand binding domain complex with methionine at 1.6 Angstrom resolution 4io7 
Crystal Structure of the AvGluR1 ligand binding domain complex with phenylalanine at 1.9 Angstrom resolution 4isu 
Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. 4iy5 
Crystal structure of the glua2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and CX516 at 2.0 A resolution 4iy6 
Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution 4jwx 
GluN2A ligand-binding core in complex with propyl-NHP5G 4jwy 
GluN2D ligand-binding core in complex with propyl-NHP5G 4kcc 
Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State 4kcd 
Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State 4kfq 
Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one 4kpt 
Crystal structure of substrate binding domain 1 (SBD1) OF ABC transporter GLNPQ from lactococcus lactis 4kqp 
Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in complex with glutamine at 0.95 A resolution 4kr5 
Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in open conformation 4l17 
GluA2-L483Y-A665C ligand-binding domain in complex with the antagonist DNQX 4la9 
Crystal structure of an empty substrate binding domain 1 (SBD1) OF ABC TRANSPORTER GLNPQ from lactococcus lactis 4lz5 
Crystal structures of GLuR2 ligand-binding-domain in complex with glutamate and positive allosteric modulators 4lz7 
Crystal structures of GLuR2 ligand-binding-domain in complex with glutamate and positive allosteric modulators 4lz8 
Crystal structures of GLuR2 ligand-binding-domain in complex with glutamate and positive allosteric modulators 4mf3 
Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist 4mh5 
Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate 4n07 
Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution 4nf4 
Crystal structure of GluN1/GluN2A ligand-binding domain in complex with DCKA and glutamate 4nf5 
Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and D-AP5 4nf6 
Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA 4nf8 
Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and glutamate in PEG2000MME 4nwc 
4NWC 4nwd 
4NWD 4o3a 
Crystal structure of the glua2 ligand-binding domain in complex with L-apartate at 1.80 a resolution 4o3b 
Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution 4o3c 
Crystal structure of the GLUA2 ligand-binding domain in complex with L-apartate at 1.50 A resolution 4oen 
Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A 4ohn 
4OHN 4owd 
4OWD 4oxv 
4OXV 4oyv 
4OYV 4oz9 
4OZ9 4p0g 
4P0G 4p0i 
4P0I 4p11 
4P11 4pow 
4POW 4pox 
4POX 4pp0 
4PP0 4prs 
4PRS 4psh 
4PSH 4q0c 
4Q0C 4q30 
4Q30 4qf9 
4QF9 4u1o 
4U1O 4u1z 
4U1Z 4u21 
4U21 4u22 
4U22 4u23 
4U23 4u2r 
4U2R 4u4s 
4U4S 4u4x 
4U4X 4wxj 
4WXJ 4x48 
4X48 4yma 
4YMA 4ymb 
4YMB 4ymx 
4YMX 4yu0 
4YU0 4z0i 
4Z0I 4z9n 
4Z9N 4zef 
4ZEF 4zv1 
4ZV1 4zv2 
4ZV2 5a5x 
5A5X 5aa1 
5AA1 5aa2 
5AA2 5aa3 
5AA3 5aa4 
5AA4 5buu 
5BUU 5cbr 
5CBR 5cbs 
5CBS 5cc2 
5CC2 5cmk 
5CMK 5cmm 
5CMM 5ddn 
5DDN 5ddx 
5DDX 5de4 
5DE4 5dex 
5DEX 5dt6 
5DT6 5dtb 
5DTB 5ehm 
5EHM 5ehs 
5EHS 5elv 
5ELV 5eyf 
5EYF 5fhm 
5FHM 5fhn 
5FHN 5fho 
5FHO 5fth 
5FTH 5fti 
5FTI 5h8f 
5H8F 5h8h 
5H8H 5h8n 
5H8N 5h8q 
5H8Q 5h8s 
5H8S 5hmt 
5HMT 5i2k 
5I2K 5i2n 
5I2N 5i56 
5I56 5i57 
5I57 5i58 
5I58 5i59 
5I59 5ikb 
5IKB 5ito 
5ITO 5itp 
5ITP 5jty 
5JTY 5kcj 
5KCJ 5kdt 
5KDT 5kuh 
5KUH 5l9m 
5L9M 5l9o 
5L9O 5lom 
5LOM 5tp9 
5TP9 5tpa 
5TPA - Links (links to other resources describing this domain)
-
PROSITE SBP_BACTERIAL_3 INTERPRO IPR001638 PFAM SBP_bac_3

