IGc2Immunoglobulin C-2 Type |
|---|
| SMART accession number: | SM00408 |
|---|---|
| Description: | - |
| Interpro abstract (IPR003598): | The basic structure of immunoglobulin (Ig) molecules is a tetramer of two light chains and two heavy chains linked by disulphide bonds. There are two types of light chains: kappa and lambda, each composed of a constant domain (CL) and a variable domain (VL). There are five types of heavy chains: alpha, delta, epsilon, gamma and mu, all consisting of a variable domain (VH) and three (in alpha, delta and gamma) or four (in epsilon and mu) constant domains (CH1 to CH4). Ig molecules are highly modular proteins, in which the variable and constant domains have clear, conserved sequence patterns. The domains in Ig and Ig-like molecules are grouped into four types: V-set (variable; IPR013106 ), C1-set (constant-1; IPR003597 ), C2-set (constant-2; IPR008424 ) and I-set (intermediate; IPR013098 ) [ (PUBMED:9417933) ]. Structural studies have shown that these domains share a common core Greek-key beta-sandwich structure, with the types differing in the number of strands in the beta-sheets as well as in their sequence patterns [ (PUBMED:15327963) (PUBMED:11377196) ]. Immunoglobulin-like domains that are related in both sequence and structure can be found in several diverse protein families. Ig-like domains are involved in a variety of functions, including cell-cell recognition, cell-surface receptors, muscle structure and the immune system [ (PUBMED:10698639) ]. This entry represents a subtype of the immunoglobulin domain, and is found in a diverse range of protein families that includes glycoproteins, fibroblast growth factor receptors, vascular endothelial growth factor receptors, interleukin-6 receptor, and neural cell adhesion molecules. It also includes proteins that are classified as unassigned proteinase inhibitors belonging to MEROPS inhibitor families I2, I17 and I43 [ (PUBMED:14705960) ]. |
| Family alignment: |
There are 363480 IGc2 domains in 114582 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing IGc2 domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with IGc2 domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing IGc2 domain in the selected taxonomic class.
- Disease (disease genes where sequence variants are found in this domain)
-
SwissProt sequences and OMIM curated human diseases associated with missense mutations within the IGc2 domain.
Protein Disease Low affinity immunoglobulin gamma Fc region receptor III-A (P08637) (SMART) OMIM:146740: {Lupus erythematosus, systemic, susceptibility}
OMIM:152700: Neutropenia, alloimmune neonatal ; {Viral infections, recurrent}Netrin receptor DCC (P43146) (SMART) OMIM:120470: Colorectal cancer Fibroblast growth factor receptor 2 (P21802) (SMART) OMIM:176943: Crouzon syndrome
OMIM:123500: Jackson-Weiss syndrome
OMIM:123150: Beare-Stevenson cutis gyrata syndrome
OMIM:123790: Pfeiffer syndrome
OMIM:101600: Apert syndrome
OMIM:101200: Saethre-Chotzen syndromeLow affinity immunoglobulin gamma Fc region receptor II-a (P12318) (SMART) OMIM:146790: {Lupus nephritis, susceptibility to} Myosin-binding protein C, cardiac-type (Q14896) (SMART) OMIM:600958: Cardiomyopathy, familial hypertrophic, 4
OMIM:115197:Neural cell adhesion molecule L1 (P32004) (SMART) OMIM:308840: Hydrocephalus due to aqueductal stenosis
OMIM:307000: MASA syndrome
OMIM:303350: Spastic paraplegia
OMIM:312900: - Metabolism (metabolic pathways involving proteins which contain this domain)
-
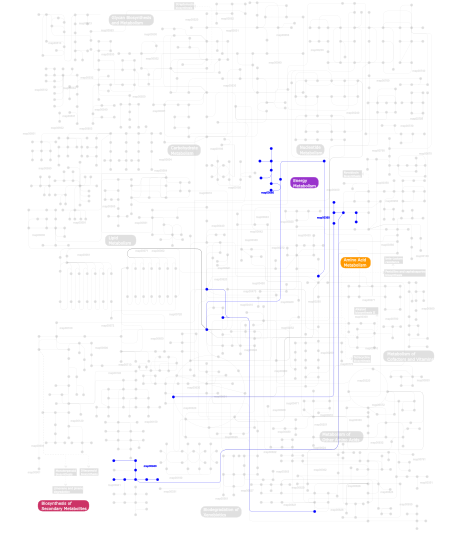
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 19.92 map04514 Cell adhesion molecules (CAMs) 9.21 map04060 Cytokine-cytokine receptor interaction 7.59 map04360 Axon guidance 7.18 map04810 Regulation of actin cytoskeleton 6.23 map04640 Hematopoietic cell lineage 6.10 map04510 Focal adhesion 4.88 map04670 Leukocyte transendothelial migration 3.79 map05215 Prostate cancer 2.98 map04520 Adherens junction 2.85 map04662 B cell receptor signaling pathway 2.85 map04660 T cell receptor signaling pathway 2.57 map05120 Epithelial cell signaling in Helicobacter pylori infection 2.57 map04020 Calcium signaling pathway 2.57 map04010 MAPK signaling pathway 2.57 map05218 Melanoma 1.90 map05210 Colorectal cancer 1.22 map04370 VEGF signaling pathway 1.22 map05214 Glioma 1.22 map04540 Gap junction 1.08 map05219 Bladder cancer 1.08 map04012 ErbB signaling pathway 0.95 map04916 Melanogenesis 0.95 map04530 Tight junction 0.95 map04620 Toll-like receptor signaling pathway 0.95 map04940 Type I diabetes mellitus 0.95 map05221 Acute myeloid leukemia 0.81 map04910 Insulin signaling pathway 0.54  map00940
map00940Phenylpropanoid biosynthesis 0.54 map04512 ECM-receptor interaction 0.54  map00680
map00680Methane metabolism 0.54 map04664 Fc epsilon RI signaling pathway 0.54  map00360
map00360Phenylalanine metabolism 0.14 map04650 Natural killer cell mediated cytotoxicity This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with IGc2 domain which could be assigned to a KEGG orthologous group, and not all proteins containing IGc2 domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of IGc2 domains in PDB
PDB code Main view Title 1bih 
CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION 1cs6 
N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN 1cvs 
CRYSTAL STRUCTURE OF A DIMERIC FGF2-FGFR1 COMPLEX 1djs 
LIGAND-BINDING PORTION OF FIBROBLAST GROWTH FACTOR RECEPTOR 2 IN COMPLEX WITH FGF1 1e07 
Model of human carcinoembryonic antigen by homology modelling and curve-fitting to experimental solution scattering data 1e0o 
Crystal structure of a ternary FGF1-FGFR2-heparin complex 1epf 
CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) 1ev2 
CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) 1evt 
CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) 1f2q 
CRYSTAL STRUCTURE OF THE HUMAN HIGH-AFFINITY IGE RECEPTOR 1f42 
THE P40 DOMAIN OF HUMAN INTERLEUKIN-12 1f45 
HUMAN INTERLEUKIN-12 1f6a 
Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) 1fhg 
HIGH RESOLUTION REFINEMENT OF TELOKIN 1fq9 
CRYSTAL STRUCTURE OF A TERNARY FGF2-FGFR1-HEPARIN COMPLEX 1gl4 
Nidogen-1 G2/Perlecan IG3 Complex 1h5b 
T cell receptor Valpha11 (AV11S5) domain 1ie5 
NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. 1ii4 
CRYSTAL STRUCTURE OF SER252TRP APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 1iil 
CRYSTAL STRUCTURE OF PRO253ARG APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 1j86 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), MONOCLINIC CRYSTAL FORM 2 1j87 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), HEXAGONAL CRYSTAL FORM 1 1j88 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 1 1j89 
HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 2 1jbj 
CD3 Epsilon and gamma Ectodomain Fragment Complex in Single-Chain Construct 1koa 
TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS 1l6z 
CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY 1n26 
Crystal Structure of the extra-cellular domains of Human Interleukin-6 Receptor alpha chain 1nct 
TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR 1ncu 
TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR 1nun 
Crystal Structure Analysis of the FGF10-FGFR2b Complex 1qz1 
Crystal Structure of the Ig 1-2-3 fragment of NCAM 1rhf 
Crystal Structure of human Tyro3-D1D2 1rpq 
High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display 1ry7 
Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 1sy6 
Crystal Structure of CD3gammaepsilon Heterodimer in Complex with OKT3 Fab Fragment 1tlk 
X-RAY STRUCTURE DETERMINATION OF TELOKIN, THE C-TERMINAL DOMAIN OF MYOSIN LIGHT CHAIN KINASE, AT 2.8 ANGSTROMS RESOLUTION 1tnm 
TERTIARY STRUCTURE OF AN IMMUNOGLOBULIN-LIKE DOMAIN FROM THE GIANT MUSCLE PROTEIN TITIN: A NEW MEMBER OF THE I SET 1tnn 
TERTIARY STRUCTURE OF AN IMMUNOGLOBULIN-LIKE DOMAIN FROM THE GIANT MUSCLE PROTEIN TITIN: A NEW MEMBER OF THE I SET 1u2h 
X-ray Structure of the N-terminally truncated human APEP-1 1wvz 
Solution Structure of the D2 Domain of the Fibroblast Growth Factor 1xiw 
Crystal structure of human CD3-e/d dimer in complex with a UCHT1 single-chain antibody fragment 1xmw 
CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT 1ya5 
Crystal structure of the titin domains z1z2 in complex with telethonin 2a38 
Crystal structure of the N-Terminus of titin 2ckn 
NMR Structure of the First Ig Module of mouse FGFR1 2cpc 
Solution structure of RSGI RUH-030, an Ig like domain from human cDNA 2cr3 
Solution structure of the first Ig-like domain of human fibroblast growth factor receptor 1 2d3v 
Crystal Structure of Leukocyte Ig-like Receptor A5 (LILRA5/LIR9/ILT11) 2dl9 
Solution structure of the Ig-like domain of human Leucine-rich repeat-containing protein 4 2dm2 
Solution structure of the first ig domain of human palladin 2dm3 
Solution structure of the second ig domain of human palladin 2dm7 
Solution structure of the 14th Ig-like domain of human KIAA1556 protein 2e9w 
Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) 2ec8 
Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit 2edf 
Solution structure of the second ig-like domain(2826-2915) from human Obscurin 2edj 
Solution structure of the fifth ig-like domain from human Roundabout homolog 2 2edl 
Solution structure of the ig-like domain (3801-3897) of human obscurin 2edw 
Solution structure of the I-set domain (3537-3630) of human obscurin 2ens 
Solution structure of the third ig-like domain from human Advanced glycosylation end product-specific receptor 2eo9 
Solution structure of the fifth ig-like domain from human Roundabout homo1 2f8v 
Structure of full length telethonin in complex with the N-terminus of titin 2fdb 
Crystal Structure of Fibroblast growth factor (FGF)8b in complex with FGF Receptor (FGFR) 2c 2id5 
Crystal Structure of the Lingo-1 Ectodomain 2iep 
Crystal structure of immunoglobulin-like domains 1 and 2 of the receptor tyrosine kinase MuSK 2jll 
Crystal structure of NCAM2 IgIV-FN3II 2kkq 
Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158. 2le9 
RAGEC2-S100A13 tetrameric complex 2lqr 
NMR structure of Ig3 domain of palladin 2ncm 
NEURAL CELL ADHESION MOLECULE, NMR, 20 STRUCTURES 2npl 
NMR Structure of CARD d2 Domain 2o26 
Structure of a class III RTK signaling assembly 2om5 
N-Terminal Fragment of Human TAX1 2r15 
Crystal structure of the myomesin domains 12 and 13 2rik 
I-band fragment I67-I69 from titin 2rjm 
3Ig structure of titin domains I67-I69 E-to-A mutated variant 2v5m 
Structural basis for Dscam isoform specificity 2v5r 
Structural basis for Dscam isoform specificity 2v5s 
Structural basis for Dscam isoform specificity 2v5t 
Crystal structure of NCAM2 Ig2-3 2v9q 
First and second Ig domains from human Robo1 2v9r 
First and second Ig domains from human Robo1 (Form 2) 2v9t 
Complex between the second LRR domain of Slit2 and The first Ig domain from Robo1 2vaj 
Crystal structure of NCAM2 Ig1 (I4122 cell unit) 2vr9 
Drosophila Robo IG1-2 (tetragonal form) 2vra 
Drosophila Robo IG1-2 (monoclinic form) 2wim 
Crystal structure of NCAM2 IG1-3 2wp3 
Crystal structure of the Titin M10-Obscurin like 1 Ig complex 2wv3 
Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor 2wwk 
Crystal structure of the Titin M10-Obscurin like 1 Ig F17R mutant complex 2wwm 
Crystal structure of the Titin M10-Obscurin like 1 Ig complex in space group P1 2xy1 
CRYSTAL STRUCTURE OF NCAM2 IG3-4 2xy2 
CRYSTAL STRUCTURE OF NCAM2 IG1-2 2xyc 
CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I 2y25 
Crystal structure of the myomesin domains My11-My13 2y7q 
The high-affinity complex between IgE and its receptor Fc epsilon RI 2yd1 
Crystal structure of the N-terminal Ig1-2 module of Drosophila Receptor Protein Tyrosine Phosphatase DLAR 2yd2 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma 2yd3 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma 2yd4 
Crystal structure of the N-terminal Ig1-2 module of Chicken Receptor Protein Tyrosine Phosphatase Sigma 2yd5 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR 2yd6 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Delta 2yd7 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Delta 2yd8 
Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR in complex with sucrose octasulphate 2yd9 
Crystal structure of the N-terminal Ig1-3 module of Human Receptor Protein Tyrosine Phosphatase Sigma 2yr3 
Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle 3b43 
I-band fragment I65-I70 from titin 3caf 
Crystal Structure of hFGFR2 D2 Domain 3cu1 
Crystal Structure of 2:2:2 FGFR2D2:FGF1:SOS complex 3d85 
Crystal structure of IL-23 in complex with neutralizing FAB 3d87 
Crystal structure of Interleukin-23 3dar 
Crystal structure of D2 domain from human FGFR2 3dmk 
Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains 3duh 
Structure of Interleukin-23 3ejj 
Structure of M-CSF bound to the first three domains of FMS 3euu 
Crystal structure of the FGFR2 D2 domain 3grw 
FGFR3 in complex with a Fab 3hmx 
Crystal structure of ustekinumab FAB/IL-12 complex 3j6n 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3j6o 
Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle 3jxa 
Immunoglobulin domains 1-4 of mouse CNTN4 3jz7 
Crystal structure of the extracellular domains of coxsackie & adenovirus receptor from mouse (mCAR) 3k0w 
Crystal structure of the tandem IG-like C2-type 2 domains of the human mucosa-associated lymphoid tissue lymphoma translocation protein 1 3kld 
PTPRG CNTN4 complex 3knb 
Crystal structure of the titin C-terminus in complex with obscurin-like 1 3kvq 
Crystal structure of VEGFR2 extracellular domain D7 3laf 
Structure of DCC, a netrin-1 receptor 3lcy 
Titin Ig tandem domains A164-A165 3mj7 
Crystal structure of the complex of JAML and Coxsackie and Adenovirus receptor, CAR 3mjg 
The structure of a platelet derived growth factor receptor complex 3mtr 
Crystal structure of the Ig5-FN1 tandem of human NCAM 3ncm 
NEURAL CELL ADHESION MOLECULE, MODULE 2, NMR, 20 STRUCTURES 3oj2 
Crystal structure of FGF1 complexed with the ectodomain of FGFR2b harboring the A172F Pfeiffer syndrome mutation 3ojm 
Crystal Structure of FGF1 complexed with the ectodomain of FGFR2b harboring P253R Apert mutation 3ojv 
Crystal Structure of FGF1 complexed with the ectodomain of FGFR1c exhibiting an ordered ligand specificity-determining betaC'-betaE loop 3p3y 
Crystal structure of neurofascin homophilic adhesion complex in space group p6522 3p40 
Crystal structure of neurofascin adhesion complex in space group p3221 3pqy 
Crystal Structure of 6218 TCR in complex with the H2Db-PA224 3puc 
Atomic resolution structure of titin domain M7 3pxh 
Tandem Ig domains of tyrosine phosphatase LAR 3pxj 
Tandem Ig repeats of Dlar 3qqn 
The retinal specific CD147 Ig0 domain: from molecular structure to biological activity 3qr2 
Wild type CD147 Ig0 domain 3qwr 
Crystal structure of IL-23 in complex with an adnectin 3r08 
Crystal structure of mouse cd3epsilon in complex with antibody 2C11 Fab 3r4d 
Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor 3rjd 
Crystal structure of Fc RI and its implication to high affinity immunoglobulin G binding 3s97 
PTPRZ CNTN1 complex 3so5 
Crystal structure of an Immunoglobulin I-set domain of Lrig3 protein (Lrig3) from MUS MUSCULUS at 1.70 A resolution 3uto 
Twitchin kinase region from C.elegans (Fn31-NL-kin-CRD-Ig26) 3v2a 
VEGFR-2/VEGF-A COMPLEX STRUCTURE 3v6b 
VEGFR-2/VEGF-E complex structure 3zyi 
NetrinG2 in complex with NGL2 3zyj 
NetrinG1 in complex with NGL1 3zyo 
Crystal structure of the N-terminal leucine rich repeats and immunoglobulin domain of netrin-G ligand-3 4c4k 
4C4K 4cbp 
4CBP 4dkd 
Crystal Structure of Human Interleukin-34 Bound to Human CSF-1R 4exp 
Structure of mouse Interleukin-34 in complex with mouse FMS 4grw 
Structure of a complex of human IL-23 with 3 Nanobodies (Llama vHHs) 4hwu 
Crystal structure of the Ig-C2 type 1 domain from mouse Fibroblast growth factor receptor 2 (FGFR2) [NYSGRC-005912] 4j23 
Low resolution crystal structure of the FGFR2D2D3/FGF1/SR128545 complex 4kc3 
Cytokine/receptor binary complex 4liq 
4LIQ 4lp5 
Crystal structure of the full-length human RAGE extracellular domain (VC1C2 fragment) 4oe8 
4OE8 4ofp 
Crystal Structure of SYG-2 D3-D4 4ofy 
Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 4og9 
4OG9 4oqt 
LINGO-1/Li81 Fab complex 4p2y 
4P2Y 4pbv 
4PBV 4pbw 
4PBW 4pbx 
4PBX 4rca 
4RCA 4u7m 
4U7M 4uow 
4UOW 4w4o 
4W4O 4wrl 
4WRL 4wrm 
4WRM 4wv1 
4WV1 4x4m 
4X4M 4x83 
4X83 4x8x 
4X8X 4x9b 
4X9B 4x9f 
4X9F 4x9g 
4X9G 4x9h 
4X9H 4x9i 
4X9I 4xb7 
4XB7 4xb8 
4XB8 4xhq 
4XHQ 4y61 
4Y61 4ybh 
4YBH 4yfc 
4YFC 4yfd 
4YFD 4yfg 
4YFG 4yh6 
4YH6 4yh7 
4YH7 4zne 
4ZNE 5aea 
5AEA 5e4i 
5E4I 5e5r 
5E5R 5e5u 
5E5U 5eo9 
5EO9 5f1d 
5F1D 5fm5 
5FM5 5ftt 
5FTT 5i99 
5I99 5k6u 
5K6U 5k6v 
5K6V 5k6w 
5K6W 5k6x 
5K6X 5k6y 
5K6Y 5k6z 
5K6Z 5k70 
5K70 - Links (links to other resources describing this domain)
-
PFAM ig INTERPRO IPR003598

