HATPase_cHistidine kinase-like ATPases |
|---|
| SMART accession number: | SM00387 |
|---|---|
| Description: | Histidine kinase-, DNA gyrase B-, phytochrome-like ATPases. |
| Interpro abstract (IPR003594): | This domain is found in several ATP-binding proteins, including: histidine kinase [ (PUBMED:15157101) ], DNA gyrase B, topoisomerases [ (PUBMED:15105144) ], heat shock protein HSP90 [ (PUBMED:15292259) (PUBMED:14718169) (PUBMED:15217611) ], phytochrome-like ATPases and DNA mismatch repair proteins. The fold of this domain consists of two layers, alpha/beta, which contains an 8-stranded mixed beta-sheet. |
| Family alignment: |
There are 925732 HATPase_c domains in 922402 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing HATPase_c domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with HATPase_c domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing HATPase_c domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Cellular role: signalling
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
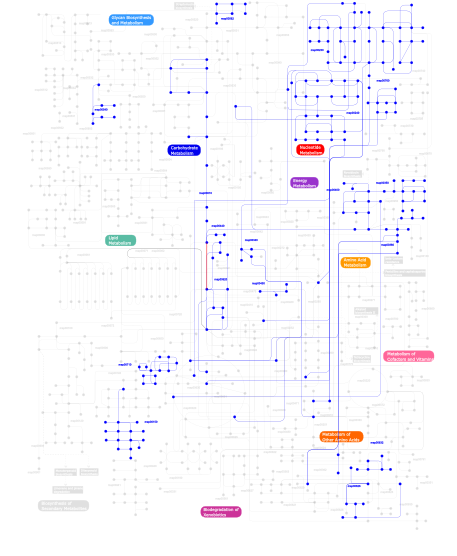
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 66.93 map02020 Two-component system - General 7.80 map02030 Bacterial chemotaxis - General 6.73 map05215 Prostate cancer 6.40 map04612 Antigen processing and presentation 6.40 map04914 Progesterone-mediated oocyte maturation 2.14 map03090 Type II secretion system 0.60 map05210 Colorectal cancer 0.60 map05213 Endometrial cancer 0.41  map00562
map00562Inositol phosphate metabolism 0.41  map00632
map00632Benzoate degradation via CoA ligation 0.25  map00540
map00540Lipopolysaccharide biosynthesis 0.22  map00230
map00230Purine metabolism 0.14  map00620
map00620Pyruvate metabolism 0.14 map04910 Insulin signaling pathway 0.14  map00010
map00010Glycolysis / Gluconeogenesis 0.14  map00710
map00710Carbon fixation 0.14 map04930 Type II diabetes mellitus 0.05 map02010 ABC transporters - General 0.03  map00350
map00350Tyrosine metabolism 0.03  map00760
map00760Nicotinate and nicotinamide metabolism 0.03  map00240
map00240Pyrimidine metabolism 0.03  map00400
map00400Phenylalanine, tyrosine and tryptophan biosynthesis 0.03  map00450
map00450Selenoamino acid metabolism 0.03 map04510 Focal adhesion 0.03  map00340
map00340Histidine metabolism 0.03 map04810 Regulation of actin cytoskeleton 0.03  map00380
map00380Tryptophan metabolism 0.03  map00440
map00440Aminophosphonate metabolism 0.03  map00626
map00626Naphthalene and anthracene degradation 0.03  map00150
map00150Androgen and estrogen metabolism 0.03 map04020 Calcium signaling pathway This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with HATPase_c domain which could be assigned to a KEGG orthologous group, and not all proteins containing HATPase_c domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of HATPase_c domains in PDB
PDB code Main view Title 1a4h 
STRUCTURE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE IN COMPLEX WITH GELDANAMYCIN 1ah6 
STRUCTURE OF THE TETRAGONAL FORM OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE 1ah8 
STRUCTURE OF THE ORTHORHOMBIC FORM OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE 1aj6 
NOVOBIOCIN-RESISTANT MUTANT (R136H) OF THE N-TERMINAL 24 KDA FRAGMENT OF DNA GYRASE B COMPLEXED WITH NOVOBIOCIN AT 2.3 ANGSTROMS RESOLUTION 1am1 
ATP BINDING SITE IN THE HSP90 MOLECULAR CHAPERONE 1amw 
ADP BINDING SITE IN THE HSP90 MOLECULAR CHAPERONE 1b3q 
CRYSTAL STRUCTURE OF CHEA-289, A SIGNAL TRANSDUCING HISTIDINE KINASE 1b62 
MUTL COMPLEXED WITH ADP 1b63 
MUTL COMPLEXED WITH ADPNP 1bgq 
RADICICOL BOUND TO THE ATP BINDING SITE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE 1bkn 
CRYSTAL STRUCTURE OF AN N-TERMINAL 40KD FRAGMENT OF E. COLI DNA MISMATCH REPAIR PROTEIN MUTL 1bxd 
NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ 1byq 
HSP90 N-TERMINAL DOMAIN BOUND TO ADP-MG 1ea6 
N-terminal 40kDa fragment of NhPMS2 complexed with ADP 1ei1 
DIMERIZATION OF E. COLI DNA GYRASE B PROVIDES A STRUCTURAL MECHANISM FOR ACTIVATING THE ATPASE CATALYTIC CENTER 1gjv 
Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) complxed with ATP-gamma-S 1gkx 
Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) 1gkz 
Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) complxed with ADP 1h7s 
N-terminal 40kDa fragment of human PMS2 1h7u 
NhPMS2-ATPgS 1i58 
STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ATP ANALOG ADPCP AND MAGNESIUM 1i59 
STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ADPNP AND MAGENSIUM 1i5a 
STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPCP AND MANGANESE 1i5b 
STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE 1i5c 
STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADP 1i5d 
STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH TNP-ATP 1id0 
CRYSTAL STRUCTURE OF THE NUCLEOTIDE BOND CONFORMATION OF PHOQ KINASE DOMAIN 1jm6 
Pyruvate dehydrogenase kinase, isozyme 2, containing ADP 1kij 
Crystal structure of the 43K ATPase domain of Thermus thermophilus gyrase B in complex with novobiocin 1kzn 
Crystal Structure of E. coli 24kDa Domain in Complex with Clorobiocin 1l0o 
Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF 1mu5 
Structure of topoisomerase subunit 1mx0 
Structure of topoisomerase subunit 1nhh 
Crystal structure of N-terminal 40KD MutL protein (LN40) complex with ADPnP and one Rubidium 1nhi 
Crystal structure of N-terminal 40KD MutL (LN40) complex with ADPnP and one potassium 1nhj 
Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium 1osf 
Human Hsp90 in complex with 17-desmethoxy-17-N,N-Dimethylaminoethylamino-Geldanamycin 1pvg 
Crystal Structure of the ATPase region of Saccharomyces Cerevisiae topoisomerase II 1qy5 
Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with the specific ligand NECA 1qy8 
Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with Radicicol 1qye 
Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with 2-chlorodideoxyadenosine 1qzr 
CRYSTAL STRUCTURE OF THE ATPASE REGION OF SACCHAROMYCES CEREVISIAE TOPOISOMERASE II BOUND TO ICRF-187 (DEXRAZOXANE) 1r62 
Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) 1s14 
Crystal structure of Escherichia coli Topoisomerase IV ParE 24kDa subunit 1s16 
Crystal Structure of E. coli Topoisomerase IV ParE 43kDa subunit complexed with ADPNP 1tbw 
Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation 1tc0 
Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP 1tc6 
Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation ADP-Complex 1th8 
Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form II 1thn 
Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I 1tid 
Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: Poised for phosphorylation complex with ATP, crystal form I 1til 
Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA:Poised for phosphorylation complex with ATP, crystal form II 1u0y 
N-Domain Of Grp94, with the Charged Domain, In Complex With the Novel Ligand N-Propyl Carboxyamido Adenosine 1u0z 
N-Domain Of Grp94 Lacking The Charged Domain In Complex With Radicicol 1u2o 
Crystal Structure Of The N-Domain Of Grp94 Lacking The Charged Domain In Complex With Neca 1us7 
Complex of Hsp90 and P50 1uy6 
Human Hsp90-alpha with 9-Butyl-8-(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine 1uy7 
Human Hsp90-alpha with 9-Butyl-8-(4-methoxy-benzyl)-9H-purin-6-ylamine 1uy8 
Human Hsp90-alpha with 9-Butyl-8-(3-trimethoxy-benzyl)-9H-purin-6ylamine 1uy9 
Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-9H-purin-6-ylamine 1uyc 
Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-9H-purin-6-ylamine 1uyd 
Human Hsp90-alpha with 9-Butyl-8-(2-chloro-3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine 1uye 
Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-9-pent-4-ylnyl-9H-purin-6-ylamine 1uyf 
Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-2-fluoro-9-pent-4-ylnyl-9H-purin-6-ylamine 1uyg 
Human Hsp90-alpha with 8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine 1uyh 
Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine 1uyi 
Human Hsp90-alpha with 8-(2,5-dimethoxy-benzyl)-2-fluoro-9-pent-9H-purin-6-ylamine 1uyk 
Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-2-fluoro-9H-purin-6-ylamine 1uyl 
Structure-Activity Relationships in purine-based inhibitor binding to HSP90 isoforms 1uym 
Human Hsp90-beta with PU3 (9-Butyl-8(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine) 1y4s 
Conformation rearrangement of heat shock protein 90 upon ADP binding 1y4u 
Conformation rearrangement of heat shock protein 90 upon ADP binding 1y8n 
Crystal structure of the PDK3-L2 complex 1y8o 
Crystal structure of the PDK3-L2 complex 1y8p 
Crystal structure of the PDK3-L2 complex 1yc1 
Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles 1yc3 
Crystal Structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles 1yc4 
Crystal structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles 1yer 
HUMAN HSP90 GELDANAMYCIN-BINDING DOMAIN, "CLOSED" CONFORMATION 1yes 
HUMAN HSP90 GELDANAMYCIN-BINDING DOMAIN, "OPEN" CONFORMATION 1yet 
GELDANAMYCIN BOUND TO THE HSP90 GELDANAMYCIN-BINDING DOMAIN 1ys3 
Crystal Structure of the ATP binding domain of PrrB from Mycobacterium Tuberculosis 1ysr 
Crystal Structure of ATP binding domain of PrrB from Mycobacterium Tuberculosis 1ysz 
Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH NECA 1yt0 
Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH ADP 1yt1 
Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL 1yt2 
Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N APO CRYSTAL 1z59 
Topoisomerase VI-B, ADP-bound monomer form 1z5a 
Topoisomerase VI-B, ADP-bound dimer form 1z5b 
Topoisomerase VI-B, ADP AlF4- bound dimer form 1z5c 
Topoisomerase VI-B, ADP Pi bound dimer form 1zw9 
Yeast HSP82 in complex with the Novel HSP90 Inhibitor 8-(6-Bromo-benzo[1,3]dioxol-5-ylsulfanyl)-9-(3-isopropylamino-propyl)-adenine 1zwh 
Yeast Hsp82 in complex with the novel Hsp90 inhibitor Radester amine 1zxm 
Human Topo IIa ATPase/AMP-PNP 1zxn 
Human DNA topoisomerase IIa ATPase/ADP 2akp 
Hsp90 Delta24-N210 mutant 2brc 
Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. 2bre 
STRUCTURE OF A HSP90 INHIBITOR BOUND TO THE N-TERMINUS OF YEAST HSP90. 2bsm 
Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design 2bt0 
Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design 2btz 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2bu2 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2bu5 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2bu6 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2bu7 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2bu8 
crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands 2byh 
3-(5-chloro-2,4-dihydroxyphenyl)-pyrazole-4-carboxamides as Inhibitors of the Hsp90 Molecular Chaperone 2byi 
3-(5-chloro-2,4-dihydroxyphenyl)-pyrazole-4-carboxamides as Inhibitors of the Hsp90 Molecular Chaperone 2bz5 
Structure-based Discovery of a New Class of Hsp90 Inhibitors 2c2a 
Structure of the entire cytoplasmic portion of a sensor histidine kinase protein 2ccs 
HUMAN HSP90 WITH 4-CHLORO-6-(4-PIPERAZIN-1-YL-1H-PYRAZOL-3-YL)- BENZENE-1,2-DIOL 2cct 
HUMAN HSP90 WITH 5-(5-CHLORO-2,4-DIHYDROXY-PHENYL)-4-PIPERAZIN-1-YL- 2H-PYRAZOLE-3-CARBOXYLIC ACID ETHYLAMIDE 2ccu 
HUMAN HSP90 WITH 4-CHLORO-6-(4-(4-(4-METHANESULPHONYL-BENZYL)- PIERAZIN-1-YL)-1H-PYRAZOL-3-YL)-BENZENE-1,3-DIOL 2cg9 
Crystal structure of an Hsp90-Sba1 closed chaperone complex 2cgf 
A RADICICOL ANALOGUE BOUND TO THE ATP BINDING SITE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE 2ch4 
Complex between Bacterial Chemotaxis histidine kinase CheA domains P4 and P5 and receptor-adaptor protein CheW 2e0a 
Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with AMPPNP 2esa 
GRP94 n-terminal domain bound to geldanamycin: effects of mutants 168-169 KS-AA 2exl 
GRP94 N-terminal Domain bound to geldanamycin 2fwy 
Structure of human Hsp90-alpha bound to the potent water soluble inhibitor PU-H64 2fwz 
Structure of human Hsp90-alpha bound to the potent water soluble inhibitor PU-H71 2fxs 
Yeast HSP82 in complex with the novel HSP90 Inhibitor Radamide 2fyp 
GRP94 in complex with the novel HSP90 Inhibitor Radester amine 2gfd 
GRP94 in complex with the novel HSP90 Inhibitor Radamide 2gqp 
N-Domain Of Grp94 In Complex With the Novel Ligand N-Propyl Carboxyamido Adenosine 2h55 
Structure of human Hsp90-alpha bound to the potent water soluble inhibitor PU-DZ8 2h8m 
N-Domain Of Grp94 In Complex With the 2-Iodo-NECA 2hch 
N-Domain Of Grp94 In Complex With the Novel Ligand N-(2-amino)ethyl Carboxyamido Adenosine 2hg1 
N-Domain Of Grp94 In Complex With the Novel Ligand N-(2-hydroxyl)ethyl Carboxyamido Adenosine 2hkj 
Topoisomerase VI-B bound to radicicol 2iop 
Crystal Structure of Full-length HtpG, the Escherichia coli Hsp90, Bound to ADP 2ioq 
Crystal Structure of full-length HTPG, the Escherichia coli HSP90 2ior 
Crystal Structure of the N-terminal Domain of HtpG, the Escherichia coli Hsp90, Bound to ADP 2iws 
Radicicol analogues bound to the ATP site of HSP90 2iwu 
Analogues of radicicol bound to the ATP-binding site of Hsp90 2iwx 
Analogues of radicicol bound to the ATP-binding site of Hsp90. 2jjc 
Hsp90 alpha ATPase domain with bound small molecule fragment 2jki 
Complex of Hsp90 N-terminal and Sgt1 CS domain 2k5b 
Human CDC37-HSP90 docking model based on NMR 2o1u 
Structure of full length GRP94 with AMP-PNP bound 2o1v 
Structure of full length GRP94 with ADP bound 2o1w 
Structure of N-terminal plus middle domains (N+M) of GRP94 2pnr 
Crystal Structure of the asymmetric Pdk3-l2 Complex 2q2e 
Crystal structure of the topoisomerase VI holoenzyme from Methanosarcina mazei 2q8f 
Structure of pyruvate dehydrogenase kinase isoform 1 2q8g 
Structure of pyruvate dehydrogenase kinase isoform 1 in complex with glucose-lowering drug AZD7545 2q8h 
Structure of pyruvate dehydrogenase kinase isoform 1 in complex with dichloroacetate (DCA) 2q8i 
Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol 2qf6 
HSP90 complexed with A56322 2qfo 
HSP90 complexed with A143571 and A516383 2qg0 
HSP90 complexed with A943037 2qg2 
HSP90 complexed with A917985 2uwd 
Inhibition of the HSP90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole, isoxazole amide analogs 2vci 
4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer 2vcj 
4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer 2vw5 
Structure Of The Hsp90 Inhibitor 7-O-carbamoylpremacbecin Bound To The N- Terminus Of Yeast Hsp90 2vwc 
STRUCTURE OF THE HSP90 INHIBITOR MACBECIN BOUND TO THE N-TERMINUS OF YEAST HSP90. 2wep 
Yeast Hsp90 N-terminal domain LI-IV mutant with ADP 2weq 
Yeast Hsp90 N-terminal domain LI-IV mutant with Geldanamycin 2wer 
Yeast Hsp90 N-terminal domain LI-IV mutant with Radicicol 2wi1 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi2 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi3 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi4 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi5 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi6 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2wi7 
Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone 2xab 
Structure of HSP90 with an inhibitor bound 2xcm 
COMPLEX OF HSP90 N-TERMINAL, SGT1 CS AND RAR1 CHORD2 DOMAIN 2xd6 
Hsp90 complexed with a resorcylic acid macrolactone. 2xdk 
Structure of HSP90 with small molecule inhibitor bound 2xdl 
Structure of HSP90 with small molecule inhibitor bound 2xds 
Structure of HSP90 with small molecule inhibitor bound 2xdu 
Structure of HSP90 with small molecule inhibitor bound 2xdx 
Structre of HSP90 with small molecule inhibitor bound 2xhr 
Structure of HSP90 with small molecule inhibitor bound 2xht 
Structure of HSP90 with small molecule inhibitor bound 2xhx 
Structure of HSP90 with small molecule inhibitor bound 2xjg 
Structure of HSP90 with small molecule inhibitor bound 2xjj 
Structure of HSP90 with small molecule inhibitor bound 2xjx 
Structure of HSP90 with small molecule inhibitor bound 2xk2 
Structure of HSP90 with small molecule inhibitor bound 2xx2 
Macrolactone Inhibitor bound to HSP90 N-term 2xx4 
Macrolactone Inhibitor bound to HSP90 N-term 2xx5 
Macrolactone Inhibitor bound to HSP90 N-term 2ye2 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye3 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye4 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye5 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye6 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye7 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye8 
HSP90 inhibitors and drugs from fragment and virtual screening 2ye9 
HSP90 inhibitors and drugs from fragment and virtual screening 2yea 
HSP90 inhibitors and drugs from fragment and virtual screening 2yeb 
HSP90 inhibitors and drugs from fragment and virtual screening 2yec 
HSP90 inhibitors and drugs from fragment and virtual screening 2yed 
HSP90 inhibitors and drugs from fragment and virtual screening 2yee 
HSP90 inhibitors and drugs from fragment and virtual screening 2yef 
HSP90 inhibitors and drugs from fragment and virtual screening 2yeg 
HSP90 inhibitors and drugs from fragment and virtual screening 2yeh 
HSP90 inhibitors and drugs from fragment and virtual screening 2yei 
HSP90 inhibitors and drugs from fragment and virtual screening 2yej 
HSP90 inhibitors and drugs from fragment and virtual screening 2yga 
E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin 2yge 
E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin 2ygf 
L89V, L93I and V136M Mutant of N-Term HSP90 complexed with Geldanamycin 2yi0 
Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. 2yi5 
Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. 2yi6 
Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. 2yi7 
Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. 2yjw 
Tricyclic series of Hsp90 inhibitors 2yjx 
Tricyclic series of Hsp90 inhibitors 2yk2 
Tricyclic series of Hsp90 inhibitors 2yk9 
Tricyclic series of Hsp90 inhibitors 2ykb 
Tricyclic series of Hsp90 inhibitors 2ykc 
Tricyclic series of Hsp90 inhibitors 2yke 
Tricyclic series of Hsp90 inhibitors 2yki 
Tricyclic series of Hsp90 inhibitors 2ykj 
Tricyclic series of Hsp90 inhibitors 2zbk 
Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms 2zdx 
Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 2zdy 
Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 2zkj 
Crystal structure of human PDK4-ADP complex 3a0r 
Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) 3a0t 
Catalytic domain of histidine kinase ThkA (TM1359) in complex with ADP and Mg ion (trigonal) 3a0w 
Catalytic domain of histidine kinase ThkA (TM1359) for MAD phasing (nucleotide free form 2, orthorombic) 3a0x 
Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 1: ammomium phosphate, monoclinic) 3a0y 
Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 3: 1,2-propanediol, orthorombic) 3a0z 
Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 4: isopropanol, orthorombic) 3b24 
Hsp90 alpha N-terminal domain in complex with an aminotriazine fragment molecule 3b25 
Hsp90 alpha N-terminal domain in complex with an inhibitor CH4675194 3b26 
Hsp90 alpha N-terminal domain in complex with an inhibitor Ro1127850 3b27 
Hsp90 alpha N-terminal domain in complex with an inhibitor Ro4919127 3b28 
Hsp90 alpha N-terminal domain in complex with an inhibitor CH5015765 3bm9 
Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 3bmy 
Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 3c0e 
Yeast Hsp82 N-terminal domain: effects of mutants 98-99 KS-AA 3c11 
Yeast Hsp82 N-terminal domain-Geldanamycin complex: effects of mutants 98-99 KS-AA 3cgy 
Crystal Structure of Salmonella Sensor Kinase PhoQ catalytic domain in complex with radicicol 3cgz 
Crystal Structure of Salmonella Sensor Kinase PhoQ catalytic domain 3crk 
Crystal structure of the PDHK2-L2 complex. 3crl 
Crystal structure of the PDHK2-L2 complex. 3d0b 
Crystal Structure of Benzamide Tetrahydro-4H-carbazol-4-one bound to Hsp90 3d2r 
Crystal structure of pyruvate dehydrogenase kinase isoform 4 in complex with ADP 3d36 
How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda 3dge 
Structure of a histidine kinase-response regulator complex reveals insights into Two-component signaling and a novel cis-autophosphorylation mechanism 3ehf 
Crystal structure of DesKC in complex with AMP-PCP 3ehg 
Crystal structure of the ATP-binding domain of DesK in complex with ATP 3ehh 
Crystal structure of DesKC-H188V in complex with ADP 3ehj 
Crystal structure of DesKC-H188V in complex with AMP-PCP 3eko 
Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone 3ekr 
Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone 3ft5 
Structure of HSP90 bound with a novel fragment 3ft8 
Structure of HSP90 bound with a noval fragment. 3fv5 
Crystal Structure of E. coli Topoisomerase IV co-complexed with inhibitor 3g75 
Crystal structure of Staphylococcus aureus Gyrase B co-complexed with 4-METHYL-5-[3-(METHYLSULFANYL)-1H-PYRAZOL-5-YL]-2-THIOPHEN-2-YL-1,3-THIAZOLE inhibitor 3g7b 
Staphylococcus aureus Gyrase B co-complex with METHYL ({5-[4-(4-HYDROXYPIPERIDIN-1-YL)-2-PHENYL-1,3-THIAZOL-5-YL]-1H-PYRAZOL-3-YL}METHYL)CARBAMATE inhibitor 3g7e 
Crystal structure of E. coli Gyrase B co-complexed with PROP-2-YN-1-YL {[5-(4-PIPERIDIN-1-YL-2-PYRIDIN-3-YL-1,3-THIAZOL-5-YL)-1H-PYRAZOL-3-YL]METHYL}CARBAMATE inhibitor 3gie 
Crystal structure of DesKC_H188E in complex with AMP-PCP 3gif 
Crystal structure of DesKC_H188E in complex with ADP 3gig 
Crystal structure of phosphorylated DesKC in complex with AMP-PCP 3h4l 
Crystal Structure of N terminal domain of a DNA repair protein 3h80 
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 3hek 
HSP90 N-terminal domain in complex with 1-{4-[(2R)-1-(5-chloro-2,4-dihydroxybenzoyl)pyrrolidin-2-yl]benzyl}-3,3-difluoropyrrolidinium 3hhu 
Human heat-shock protein 90 (HSP90) in complex with {4-[3-(2,4-dihydroxy-5-isopropyl-phenyl)-5-thioxo- 1,5-dihydro-[1,2,4]triazol-4-yl]-benzyl}-carbamic acid ethyl ester {ZK 2819} 3hyy 
Crystal structure of Hsp90 with fragment 37-D04 3hyz 
Crystal structure of Hsp90 with fragment 42-C03 3hz1 
Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 3hz5 
Crystal structure of Hsp90 with fragment Z064 3ied 
Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN 3inw 
HSP90 N-TERMINAL DOMAIN with pochoxime A 3inx 
HSP90 N-TERMINAL DOMAIN with pochoxime B 3ja6 
3JA6 3jz3 
Structure of the cytoplasmic segment of histidine kinase QseC 3k60 
Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF07_0029) bound to ADP 3k97 
HSP90 N-terminal domain in complex with 4-chloro-6-{[(2R)-2-(2-methylphenyl)pyrrolidin-1-yl]carbonyl}benzene-1,3-diol 3k98 
HSP90 N-terminal domain in complex with (1R)-2-(5-chloro-2,4-dihydroxybenzoyl)-N-ethylisoindoline-1-carboxamide 3k99 
HSP90 N-terminal domain in complex with 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol 3ke6 
The crystal structure of the RsbU and RsbW domains of Rv1364c from Mycobacterium tuberculosis 3lnu 
Crystal structure of ParE subunit 3lps 
Crystal structure of parE 3mnr 
Crystal Structure of Benzamide SNX-1321 bound to Hsp90 3nmq 
Hsp90b N-terminal domain in complex with EC44, a pyrrolo-pyrimidine methoxypyridine inhibitor 3o0i 
Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor PU-H54 3o2f 
Structure of the N-domain of GRP94 bound to the HSP90 inhibitor PU-H54 3o6o 
Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 3omu 
Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a thienopyrimidine derivative 3opd 
Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a benzamide derivative 3ow6 
Crystal Structure of HSP90 with N-Aryl-benzimidazolone I 3owb 
Crystal Structure of HSP90 with VER-49009 3owd 
Crystal Structure of HSP90 with N-Aryl-benzimidazolone II 3peh 
Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative 3pej 
Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of Macbecin 3q5j 
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin 3q5k 
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor 3q5l 
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin 3qdd 
HSP90A N-terminal domain in complex with BIIB021 3qtf 
Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity 3r4m 
Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide 3r4n 
Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide 3r4o 
Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide 3r4p 
Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide 3r91 
Macrocyclic lactams as potent Hsp90 inhibitors with excellent tumor exposure and extended biomarker activity. 3r92 
Discovery of a macrocyclic o-aminobenzamide Hsp90 inhibitor with heterocyclic tether that shows extended biomarker activity and in vivo efficacy in a mouse xenograft model. 3rkz 
Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. 3rlp 
Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine 3rlq 
Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile 3rlr 
Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile 3sl2 
ATP Forms a Stable Complex with the Essential Histidine Kinase WalK (YycG) Domain 3t0h 
Structure insights into mechanisms of ATP hydrolysis and the activation of human Hsp90 3t0z 
Hsp90 N-terminal domain bound to ATP 3t10 
HSP90 N-terminal domain bound to ACP 3t1k 
HSP90 N-terminal domain bound to ANP 3t2s 
HSP90 N-terminal domain bound to AGS 3ttz 
Crystal structure of a topoisomerase ATPase inhibitor 3tuh 
Crystal Structure of the N-terminal domain of an HSP90 in the presence of an the inhibitor ganetespib 3tz0 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex 3tz2 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex 3tz4 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex with ADP 3tz5 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex with ADP 3u2d 
S. aureus GyrB ATPase domain in complex with small molecule inhibitor 3u2k 
S. aureus GyrB ATPase domain in complex with a small molecule inhibitor 3u67 
Crystal structure of the N-terminal domain of Hsp90 from Leishmania major(LmjF33.0312)in complex with ADP 3ur1 
The structure of a ternary complex between CheA domains P4 and P5 with CheW and with a truncated fragment of TM14, a chemoreceptor analog from Thermotoga maritima. 3vad 
Crystal structure of I170F mutant branched-chain alpha-ketoacid dehydrogenase kinase in complex with 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid 3vha 
Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor 3vhc 
Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor 3vhd 
Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor, CH5164840 3wha 
Hsp90 alpha N-terminal domain in complex with a tricyclic inhibitor 3wq9 
3WQ9 3zkb 
CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP 3zkd 
CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP 3zm7 
CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPCP 3zxo 
CRYSTAL STRUCTURE OF THE MUTANT ATP-BINDING DOMAIN OF MYCOBACTERIUM TUBERCULOSIS DOSS 3zxq 
CRYSTAL STRUCTURE OF THE ATP-BINDING DOMAIN OF MYCOBACTERIUM TUBERCULOSIS DOST 4as9 
The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 4asa 
The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 4asb 
The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 4asf 
The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 4asg 
The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 4awo 
Complex of HSP90 ATPase domain with tropane derived inhibitors 4awp 
Complex of HSP90 ATPase domain with tropane derived inhibitors 4awq 
Complex of HSP90 ATPase domain with tropane derived inhibitors 4b6c 
Structure of the M. smegmatis GyrB ATPase domain in complex with an aminopyrazinamide 4b7p 
Structure of HSP90 with NMS-E973 inhibitor bound 4bae 
Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis 4biu 
Crystal structure of CpxAHDC (orthorhombic form 1) 4biv 
Crystal structure of CpxAHDC (trigonal form) 4biw 
Crystal structure of CpxAHDC (hexagonal form) 4bix 
Crystal structure of CpxAHDC (monoclinic form 1) 4biy 
Crystal structure of CpxAHDC (monoclinic form 2) 4biz 
Crystal structure of CpxAHDC (orthorhombic form 2) 4bqg 
structure of HSP90 with an inhibitor bound 4bqj 
structure of HSP90 with an inhibitor bound 4bxi 
4BXI 4cb0 
Crystal structure of CpxAHDC in complex with ATP (hexagonal form) 4ce1 
Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. 4ce2 
Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. 4ce3 
Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. 4cti 
Escherichia coli EnvZ histidine kinase catalytic part fused to Archaeoglobus fulgidus Af1503 HAMP domain 4cwf 
4CWF 4cwn 
4CWN 4cwo 
4CWO 4cwp 
4CWP 4cwq 
4CWQ 4cwr 
4CWR 4cws 
4CWS 4cwt 
4CWT 4duh 
Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor 4dzy 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with ADP 4e00 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP 4e01 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with AMPPNP 4e02 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP 4eeh 
Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-(4-Hydroxy-phenyl)-1H-indazol-6-ol 4eft 
Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-Cyclohexyl-2-(6-hydroxy-1H-indazol-3-yl)-propionitrile 4efu 
Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid benzyl-methyl-amide 4egh 
Hsp90-alpha ATPase domain in complex with (4-Hydroxyphenyl)morpholin-4-yl methanone 4egi 
Hsp90-alpha ATPase domain in complex with 2-Amino-4-ethylthio-6-methyl-1,3,5-triazine 4egk 
Human Hsp90-alpha ATPase domain bound to Radicicol 4em7 
Crystal structure of a topoisomerase ATP inhibitor 4emv 
Crystal structure of a topoisomerase ATP inhibitor 4fcp 
Targetting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo [2,3-d] pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization 4fcq 
Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization 4fcr 
Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization 4gcz 
Structure of a blue-light photoreceptor 4gee 
Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4gfh 
Topoisomerase II-DNA-AMPPNP complex 4gfn 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic 4ggl 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity 4gqt 
N-terminal domain of C. elegans Hsp90 4gt8 
Crystal Structure of the Catalytic and ATP-binding Domain from VraS in Complex with ADP 4h7q 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase in complex with alpha-ketoisocaproic acid and ADP 4h81 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-2-chloro-3-phenylpropanoic acid complex with ADP 4h85 
Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-alpha-chloroisocaproate complex with ADP 4hxw 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hxz 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hy1 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hy6 
Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 4hym 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hyp 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hz0 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. 4hz5 
Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity 4i3h 
A three-gate structure of topoisomerase IV from Streptococcus pneumoniae 4i5s 
Structure and function of sensor histidine kinase 4ipe 
Crystal structure of mitochondrial Hsp90 (TRAP1) with AMPPNP 4ivg 
Crystal structure of mitochondrial Hsp90 (TRAP1) NTD-Middle domain dimer with AMPPNP 4iyn 
Structure of mitochondrial Hsp90 (TRAP1) with ADP-ALF4- 4j0b 
Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 4jas 
Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) 4jau 
Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E) 4jav 
Structural basis of a rationally rewired protein-protein interface (HK853wt and RR468mutant V13P, L14I, I17M and N21V) 4jpb 
The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. 4jql 
Synthesis of Benzoquinone-Ansamycin-Inspired Macrocyclic Lactams from Shikimic Acid 4juo 
A low-resolution three-gate structure of topoisomerase IV from Streptococcus pneumoniae in space group H32 4k4o 
The DNA Gyrase B ATP binding domain of Enterococcus faecalis in complex with a small molecule inhibitor 4kfg 
The DNA Gyrase B ATP binding domain of Escherichia coli in complex with a small molecule inhibitor. 4kp4 
Deciphering cis-trans directionality and visualizing autophosphorylation in histidine kinases. 4kqv 
Topoisomerase iv atp binding domain of francisella tularensis in complex with a small molecule inhibitor 4ksg 
Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) 4ksh 
Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (7-({4-[(3R)-3-AMINOPYRROLIDIN-1-YL]-5-CHLORO-6-ETHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-2-YL}SULFANYL)-1,5-NAPHTHYRIDIN-1(4H)-OL) 4ktn 
Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor ((3S)-1-[2-(PYRIDO[2,3-B]PYRAZIN-7-YLSULFANYL)-9H-PYRIMIDO[4,5-B]INDOL-4-YL]PYRROLIDIN-3-AMINE) 4l8z 
4L8Z 4l90 
4L90 4l91 
4L91 4l93 
4L93 4l94 
4L94 4lp0 
Crystal structure of a topoisomerase ATP inhibitor 4lpb 
Crystal structure of a topoisomerase ATPase inhibitor 4lwe 
4LWE 4lwf 
4LWF 4lwg 
4LWG 4lwh 
4LWH 4lwi 
4LWI 4mb9 
Structure of Streptococcus pneumonia ParE in complex with AZ13102335 4mbc 
Structure of Streptococcus pneumonia ParE in complex with AZ13053807 4mot 
Structure of Streptococcus pneumonia pare in complex with AZ13072886 4mp2 
Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PA1 4mp7 
Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PA7 4mpc 
Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS2 4mpe 
Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS8 4mpn 
Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS10 4nh7 
Correlation between chemotype-dependent binding conformations of HSP90 alpha/beta and isoform selectivity 4nh8 
Correlation between chemotype-dependent binding conformations of HSP90 alpha/beta and isoform selectivity 4nh9 
Correlation between chemotype-dependent binding conformations of HSP90 alpha/beta and isoform selectivity 4o04 
4O04 4o05 
Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease 4o07 
Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease 4o09 
Identification of novel HSP90 / isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington s disease 4o0b 
Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease 4p7a 
Crystal Structure of human MLH1 4p8o 
4P8O 4pl9 
4PL9 4prv 
4PRV 4prx 
4PRX 4pu9 
4PU9 4q20 
Crystal structure of a C-terminal part of tyrosine kinase (DivL) from Caulobacter crescentus CB15 at 2.50 A resolution (PSI Community Target, Shapiro) 4r1f 
4R1F 4r39 
4R39 4r3a 
4R3A 4r3m 
4R3M 4u7n 
4U7N 4u7o 
4U7O 4u93 
4U93 4url 
4URL 4urm 
4URM 4urn 
4URN 4uro 
4URO 4v25 
4V25 4v26 
4V26 4w7t 
4W7T 4wub 
4WUB 4wuc 
4WUC 4wud 
4WUD 4x9l 
4X9L 4xc0 
4XC0 4xcj 
4XCJ 4xcl 
4XCL 4xd8 
4XD8 4xdm 
4XDM 4xe2 
4XE2 4xip 
4XIP 4xiq 
4XIQ 4xir 
4XIR 4xit 
4XIT 4xiv 
4XIV 4xka 
4XKA 4xkk 
4XKK 4xko 
4XKO 4xtj 
4XTJ 4ykq 
4YKQ 4ykr 
4YKR 4ykt 
4YKT 4yku 
4YKU 4ykw 
4YKW 4ykx 
4YKX 4yky 
4YKY 4ykz 
4YKZ 4z1f 
4Z1F 4z1g 
4Z1G 4z1h 
4Z1H 4z1i 
4Z1I 4zki 
4ZKI 4zvi 
4ZVI 5akb 
5AKB 5akc 
5AKC 5akd 
5AKD 5boc 
5BOC 5bod 
5BOD 5c93 
5C93 5cf0 
5CF0 5cph 
5CPH 5ctu 
5CTU 5ctw 
5CTW 5ctx 
5CTX 5cty 
5CTY 5d6p 
5D6P 5d6q 
5D6Q 5d7c 
5D7C 5d7d 
5D7D 5d7r 
5D7R 5f3k 
5F3K 5f5r 
5F5R 5fnc 
5FNC 5fnd 
5FND 5fnf 
5FNF 5fwk 
5FWK 5fwl 
5FWL 5fwm 
5FWM 5fwp 
5FWP 5hph 
5HPH 5idj 
5IDJ 5idm 
5IDM 5in9 
5IN9 5l3j 
5L3J - Links (links to other resources describing this domain)
-
INTERPRO IPR003594

