The first of these (Figure 1a) presents a schematic of the domains identified in your query sequence (drawn approximately to scale).
Due to possible overlapping predictions (a persistent problem in automatic annotation methods) there may be alternative domain representations shown (Figure 1b).
If your sequence is predicted to contain none of the domains present in the SMART domain database then you are presented with a "Sorry, your sequence contains none of the domains searched for by SMART !!" statement.
The second line underneath the domain architecture schematic represents the result of predictions of coils (using Coils2), transmembrane regions (TopPred), signal peptides (SignalP) and regions of low compositional complexity (SEG) (figure 2).
Figure 1a
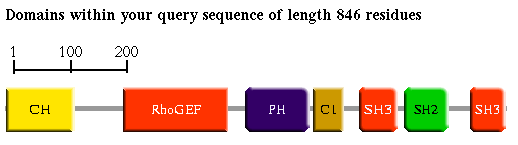
Figure 1b

Clicking on a domain icon (e.g. CH, RhoGEF, PH, ... as shown above), reveals an annotation page with special features.
In addition to information relevant to the structure and function of the domain (via links to Medline, MMDB and Prosite entries), the user may request the domain segment sequence itself, an alignment of family members, a consensus sequence of the family, the alignment of the domain segment with the profile, or else initiate a WU-Blast2 search with the domain found.
The horizontal grey bars separating the domains (the SignalP/COILS/Transmembrane/SEG hits res.) represent the regions of your query sequence without significance for the search specified.
Clicking at these 'separators' invokes a form to start a Blast2 search (note: not clickable if the length is less than 25 AA).
Clickable SIGNAL/COILS/TRANS/SEG Segments
Predictions from the SignalP, Coils2, TopPred and Seg programs are clickable also. The corresponding sequences are displayed and WU-BLAST2 requests may be initiated.
Figure 2
