Clicking on any of the SMART domain icons displayed in output pages will lead to display of a second type of annotation page.
The output is similar to figure 1 presented below.
Figure 1
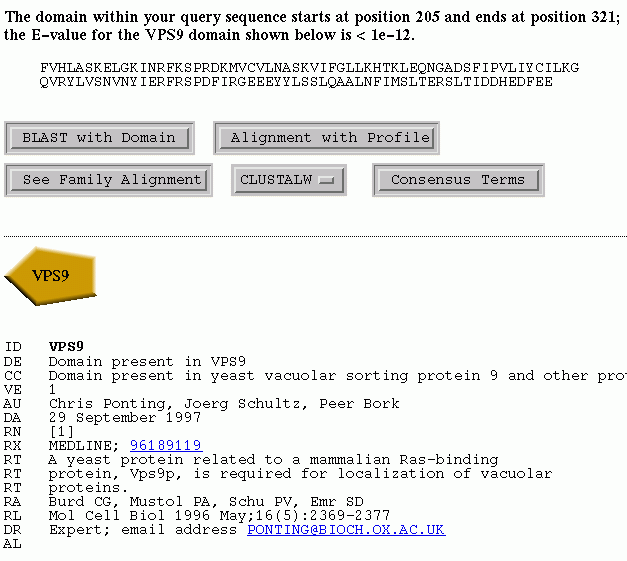
In addition to comment lines, we present links to relevant Medline, MMDB and Prosite entries.
There are 5 services that may be requested from these pages:
This initiates a Blast2 search using the domain segment as the query sequence.
As a default the search is of the "non-redundant protein database" (nrdb)
Align the domain segment sequence to the profile
his initiates an alignment of the domain segment sequence with the domain
profile using the WiseTools program, PairWise.
The alignment is shown on one or more lines with identities indicated by an asterisk "*" (see the Figure below).
This alignment is decorated with annotations that are retrieved automatically from SwissProt entries. In particular the secondary structures of domains are presented (E=beta strand, H=helix, T=turn) under the alignment, and features such as binding, catalytic, amino acid substitution, phosphorylation, and glycosylation sites are numbered according to their position above the alignment.
A key that follows the alignment describes these features, and allows direct access to their corresponding SwissProt entries.

View the consensus sequence of the family
This displays consensus sequences using thresholds of 80%, 65% and 50%, and amino acid groupings that are displayed elsewhere.
View the domain family alignment using different formats
This allows display of the full seed alignment of the domain family.
Retrieve annotations of other domains
Currently annotated domains are listed at the foot of each page and access to their annotation pages (of type 1) are provided by hyperlinks."