POLAcDNA polymerase A domain |
|---|
| SMART accession number: | SM00482 |
|---|---|
| Description: | - |
| Interpro abstract (IPR001098): | Synonym(s): DNA nucleotidyltransferase (DNA-directed) DNA-directed DNA polymerases( EC 2.7.7.7 ) are the key enzymes catalysing the accurate replication of DNA. They require either a small RNA molecule or a protein as a primer for the de novo synthesis of a DNA chain. A number of polymerases belong to this family [ (PUBMED:2196557) (PUBMED:1870963) (PUBMED:8451181) ]. |
| GO process: | DNA replication (GO:0006260) |
| GO function: | DNA binding (GO:0003677), DNA-directed DNA polymerase activity (GO:0003887) |
| Family alignment: |
There are 28198 POLAc domains in 28190 proteins in SMART's nrdb database.
Click on the following links for more information.
- Evolution (species in which this domain is found)
-
Taxonomic distribution of proteins containing POLAc domain.
This tree includes only several representative species. The complete taxonomic breakdown of all proteins with POLAc domain is also avaliable.
Click on the protein counts, or double click on taxonomic names to display all proteins containing POLAc domain in the selected taxonomic class.
- Cellular role (predicted cellular role)
-
Cellular role: replication
- Literature (relevant references for this domain)
-
Primary literature is listed below; Automatically-derived, secondary literature is also avaliable.
- Braithwaite DK, Ito J
- Compilation, alignment, and phylogenetic relationships of DNA polymerases.
- Nucleic Acids Res. 1993; 21: 787-802
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA
- Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP.
- Nature. 1985; 313: 762-6
- Display abstract
The 3.3-A resolution crystal structure of the large proteolytic fragment of Escherichia coli DNA polymerase I complexed with deoxythymidine monophosphate consists of two domains, the smaller of which binds zinc-deoxythymidine monophosphate. The most striking feature of the larger domain is a deep crevice of the appropriate size and shape for binding double-stranded B-DNA. A flexible subdomain may allow the enzyme to surround completely the DNA substrate, thereby allowing processive nucleotide polymerization without enzyme dissociation.
- Metabolism (metabolic pathways involving proteins which contain this domain)
-
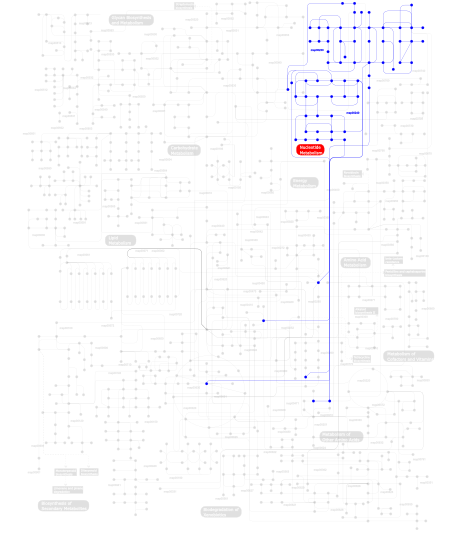
Click the image to view the interactive version of the map in iPath% proteins involved KEGG pathway ID Description 33.33  map00240
map00240Pyrimidine metabolism 33.33 map03030 DNA replication 33.33  map00230
map00230Purine metabolism This information is based on mapping of SMART genomic protein database to KEGG orthologous groups. Percentage points are related to the number of proteins with POLAc domain which could be assigned to a KEGG orthologous group, and not all proteins containing POLAc domain. Please note that proteins can be included in multiple pathways, ie. the numbers above will not always add up to 100%.
- Structure (3D structures containing this domain)
3D Structures of POLAc domains in PDB
PDB code Main view Title 1bgx 
TAQ POLYMERASE IN COMPLEX WITH TP7, AN INHIBITORY FAB 1d8y 
CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA 1d9d 
CRYSTALL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH SHORT DNA FRAGMENT CARRYING 2'-0-AMINOPROPYL-RNA MODIFICATIONS 5'-D(TCG)-AP(AUC)-3' 1d9f 
CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA TETRAMER CARRYING 2'-O-(3-AMINOPROPYL)-RNA MODIFICATION 5'-D(TT)-AP(U)-D(T)-3' 1dpi 
STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP 1jxe 
STOFFEL FRAGMENT OF TAQ DNA POLYMERASE I 1kfd 
CRYSTAL STRUCTURES OF THE KLENOW FRAGMENT OF DNA POLYMERASE I COMPLEXED WITH DEOXYNUCLEOSIDE TRIPHOSPHATE AND PYROPHOSPHATE 1kfs 
DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX 1kln 
DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX 1krp 
DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX 1ksp 
DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX 1ktq 
DNA POLYMERASE 1l3s 
Crystal Structure of Bacillus DNA Polymerase I Fragment complexed to 9 base pairs of duplex DNA. 1l3t 
Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue 1l3u 
Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue. 1l3v 
Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 15 base pairs of duplex DNA following addition of dTTP, dATP, dCTP, and dGTP residues. 1l5u 
Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 12 base pairs of duplex DNA following addition of a dTTP, a dATP, and a dCTP residue. 1lv5 
Crystal Structure of the Closed Conformation of Bacillus DNA Polymerase I Fragment Bound to DNA and dCTP 1njw 
GUANINE-THYMINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1njx 
THYMINE-GUANINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1njy 
THYMINE-THYMINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1njz 
CYTOSINE-THYMINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk0 
ADENINE-GUANINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk4 
GUANINE-GUANINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk5 
ADENINE-ADENINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk6 
CYTOSINE-CYTOSINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk7 
GUANINE-ADENINE MISMATCH AT THE POLYMERASE ACTIVE SITE 1nk8 
A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. 1nk9 
A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. 1nkb 
A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER THREE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, AND DTTP. 1nkc 
A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER FIVE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, DTTP, AND DATP. 1nke 
A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A CYTOSINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. 1qsl 
KLENOW FRAGMENT COMPLEXED WITH SINGLE-STRANDED SUBSTRATE AND EUROPIUM (III) ION 1qss 
DDGTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS 1qsy 
DDATP-Trapped closed ternary complex of the large fragment of DNA Polymerase I from thermus aquaticus 1qtm 
DDTTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS 1skr 
T7 DNA Polymerase Complexed To DNA Primer/Template and ddATP 1sks 
Binary 3' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template 1skw 
Binary 3' complex of T7 DNA polymerase with a DNA primer/template containing a disordered cis-syn thymine dimer on the template 1sl0 
Ternary 3' complex of T7 DNA polymerase with a DNA primer/template containing a disordered cis-syn thymine dimer on the template and an incoming nucleotide 1sl1 
Binary 5' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template 1sl2 
Ternary 5' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template and an incoming nucleotide 1t7p 
T7 DNA POLYMERASE COMPLEXED TO DNA PRIMER/TEMPLATE,A NUCLEOSIDE TRIPHOSPHATE, AND ITS PROCESSIVITY FACTOR THIOREDOXIN 1t8e 
T7 DNA Polymerase Ternary Complex with dCTP at the Insertion Site. 1taq 
STRUCTURE OF TAQ DNA POLYMERASE 1tau 
TAQ POLYMERASE (E.C.2.7.7.7)/DNA/B-OCTYLGLUCOSIDE COMPLEX 1tk0 
T7 DNA polymerase ternary complex with 8 oxo guanosine and ddCTP at the insertion site 1tk5 
T7 DNA polymerase binary complex with 8 oxo guanosine in the templating strand 1tk8 
T7 DNA polymerase ternary complex with 8 oxo guanosine and dAMP at the elongation site 1tkd 
T7 DNA polymerase ternary complex with 8 oxo guanosine and dCMP at the elongation site 1u45 
8oxoguanine at the pre-insertion site of the polymerase active site 1u47 
cytosine-8-Oxoguanine base pair at the polymerase active site 1u48 
Extension of a cytosine-8-oxoguanine base pair 1u49 
Adenine-8oxoguanine mismatch at the polymerase active site 1u4b 
Extension of an adenine-8oxoguanine mismatch 1ua0 
Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase 1ua1 
Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. 1x9m 
T7 DNA polymerase in complex with an N-2-acetylaminofluorene-adducted DNA 1x9s 
T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-CTP as the incoming nucleotide. 1x9w 
T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-ATP as the incoming nucleotide. 1xc9 
Structure of a high-fidelity polymerase bound to a benzo[a]pyrene adduct that blocks replication 1xwl 
BACILLUS STEAROTHERMOPHILUS (NEWLY IDENTIFIED STRAIN AS YET UNNAMED) DNA POLYMERASE FRAGMENT 1zyq 
T7 DNA polymerase in complex with 8oG and incoming ddATP 2ajq 
Structure of replicative DNA polymerase provides insigts into the mechanisms for processivity, frameshifting and editing 2bdp 
CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 9 BASE PAIRS OF DUPLEX DNA 2hhq 
O6-methyl-guanine:T pair in the polymerase-10 basepair position 2hhs 
O6-methyl:C pair in the polymerase-10 basepair position 2hht 
C:O6-methyl-guanine pair in the polymerase-2 basepair position 2hhu 
C:O6-methyl-guanine in the polymerase postinsertion site (-1 basepair position) 2hhv 
T:O6-methyl-guanine in the polymerase-2 basepair position 2hhw 
ddTTP:O6-methyl-guanine pair in the polymerase active site, in the closed conformation 2hhx 
O6-methyl-guanine in the polymerase template preinsertion site 2hvh 
ddCTP:O6MeG pair in the polymerase active site (0 position) 2hvi 
ddCTP:G pair in the polymerase active site (0 position) 2hw3 
T:O6-methyl-guanine pair in the polymerase postinsertion site (-1 basepair position) 2kfn 
KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE 2kfz 
KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND ZINC ONLY 2ktq 
OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS 2kzm 
KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE 2kzz 
KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY 2xo7 
Crystal structure of a dA:O-allylhydroxylamine-dC basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus 2xy5 
Crystal structure of an artificial salen-copper basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus 2xy6 
Crystal structure of a salicylic aldehyde basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus 2xy7 
Crystal structure of a salicylic aldehyde base in the pre-insertion site of fragment DNA polymerase I from Bacillus stearothermophilus 2y1i 
Crystal structure of a S-diastereomer analogue of the spore photoproduct in complex with fragment DNA polymerase I from Bacillus stearothermophilus 2y1j 
Crystal structure of a R-diastereomer analogue of the spore photoproduct in complex with fragment DNA polymerase I from Bacillus stearothermophilus 3bdp 
DNA POLYMERASE I/DNA COMPLEX 3eyz 
Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA (open form) 3ez5 
Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA , dCTP, and zinc (closed form). 3hp6 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus F710Y mutant bound to G:T mismatch 3hpo 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus Y714S mutant bound to G:T mismatch 3ht3 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus V713P mutant bound to G:dCTP 3ikm 
Crystal structure of human mitochondrial DNA polymerase holoenzyme 3ktq 
CRYSTAL STRUCTURE OF AN ACTIVE TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS 3lwl 
Structure of Klenow fragment of Taq polymerase in complex with an abasic site 3lwm 
Structure of the large fragment of thermus aquaticus DNA polymerase I in complex with a blunt-ended DNA and ddATP 3m8r 
Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-ethylated dTTP 3m8s 
Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-methylated dTTP 3ojs 
Snapshots of the large fragment of DNA polymerase I from Thermus Aquaticus processing C5 modified thymidines 3oju 
Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing c5 modified thymidies 3po4 
Structure of a mutant of the large fragment of DNA polymerase I from Thermus aquaticus in complex with a blunt-ended DNA and ddATP 3po5 
Structure of a mutant of the large fragment of DNA polymerase I from Thermus Auqaticus in complex with an abasic site and ddATP 3pv8 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and ddTTP-dA in Closed Conformation 3px0 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and dCTP-dA Mismatch (tautomer) in Closed Conformation 3px4 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and ddCTP-dA Mismatch (wobble) in Ajar Conformation 3px6 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and ddCTP-dA Mismatch (tautomer) in Closed Conformation 3py8 
Crystal structure of a mutant of the large fragment of DNA polymerase I from thermus aquaticus in a closed ternary complex with DNA and ddCTP 3rr7 
Binary Structure of the large fragment of Taq DNA polymerase bound to an abasic site 3rr8 
Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddGTP 3rrg 
Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddGTP 3rrh 
Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddTTP 3rtv 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with natural primer/template DNA 3sv3 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dNaM-d5SICSTP 3sv4 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase 3syz 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dNaM as templating nucleobase 3sz2 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase 3t3f 
Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and dNITP 3tan 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to Duplex DNA with Cytosine-Adenine Mismatch at (n-1) Position 3tap 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to Duplex DNA with Cytosine-Adenine Mismatch at (n-3) Position 3taq 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to Duplex DNA with Cytosine-Adenine Mismatch at (n-4) Position 3tar 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to Duplex DNA with Cytosine-Adenine Mismatch at (n-6) Position 3thv 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and ddATP-dT in Closed Conformation 3ti0 
Crystal Structure of Bacillus DNA Polymerase I Large Fragment Bound to DNA and ddGTP-dC in Closed Conformation 4b9l 
Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. 4b9m 
Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dA DNA lesion -thymine basepair in the post- insertion site. 4b9n 
Structure of the high fidelity DNA polymerase I correctly bypassing the oxidative formamidopyrimidine-dA DNA lesion. 4b9s 
Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. 4b9t 
Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dC basepair in the post-insertion site. 4b9u 
Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. 4b9v 
Structure of the high fidelity DNA polymerase I with extending from an oxidative formamidopyrimidine-dG DNA lesion -dA basepair. 4bdp 
CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 11 BASE PAIRS OF DUPLEX DNA AFTER ADDITION OF TWO DATP RESIDUES 4bwj 
KlenTaq mutant in complex with DNA and ddCTP 4bwm 
KlenTaq mutant in complex with a RNA/DNA hybrid 4c8k 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a partially closed complex with the artificial base pair d5SICS-dNaMTP 4c8l 
Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) 4c8m 
Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair d5SICS-dNaM at the postinsertion site (sequence context 2) 4c8n 
Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 3) 4c8o 
Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) 4cch 
Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide 4df4 
Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 7-(N-(10-hydroxydecanoyl)-aminopentinyl)-7-deaza-2 -dATP 4df8 
Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with aminopentinyl-7-deaza-2-dATP 4dfj 
Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 5-(aminopentinyl)-dTTP 4dfk 
large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 5-(N-(10-hydroxydecanoyl)-aminopentinyl)-2-dUTP 4dfm 
Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in ternary complex with 5-(aminopentinyl)-2-dCTP 4dfp 
Crystal structure of the large fragment of DNA Polymerase I from Thermus aqauticus in a ternary complex with 7-(aminopentinyl)-7-deaza-dGTP 4dle 
Ternary Structure of the large Fragment of Taq DNA Polymerase: 4-Fluoroproline Variant 4dlg 
Ternary Structure of the large Fragment of Taq DNA polymerase 4dqi 
Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and dCTP (paired with dG of template) 4dqp 
Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and ddCTP (paired with dG of template) 4dqq 
Ternary complex of Bacillus DNA Polymerase I Large Fragment E658A, DNA duplex, and rCTP (paired with dG of template) in presence of Mg2+ 4dqr 
Ternary complex of Bacillus DNA Polymerase I Large Fragment E658A, DNA duplex, and rCTP (paired with dG of template) in presence of Mn2+ 4dqs 
Binary complex of Bacillus DNA Polymerase I Large Fragment and duplex DNA with rC in primer terminus paired with dG of template 4ds4 
Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and rCTP in presence of Mn2+ 4ds5 
Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and rCTP in presence of Mg2+ 4dse 
Ternary complex of Bacillus DNA Polymerase I Large Fragment F710Y, DNA duplex, and rCTP (paired with dG of template) in presence of Mg2+ 4dsf 
Ternary complex of Bacillus DNA Polymerase I Large Fragment F710Y, DNA duplex, and rCTP (paired with dG of template) in presence of Mn2+ 4dsi 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, Se-dGTP and Calcium 4dsj 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, dGTP and Calcium 4dsk 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, PPi and Calcium 4dsl 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA and Calcium 4dwi 
Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with self complementary DNA, Se-dGTP and Calcium 4e0d 
Binary complex of Bacillus DNA Polymerase I Large Fragment E658A and duplex DNA 4elt 
Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines 4elu 
Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines 4elv 
Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines 4ez6 
Bacillus DNA Polymerase I Large Fragment Complex 1 4ez9 
Bacillus DNA Polymerase I Large Fragment Complex 2 4f2r 
DNA Polymerase I Large Fragment complex 3 4f2s 
DNA Polymerase I Large Fragment complex 4 4f3o 
DNA Polymerase I Large Fragment Complex 5 4f4k 
DNA Polymerase I Large Fragment Complex 6 4f8r 
Bacillus DNA Polymerase I Large Fragment complex 7 4ktq 
BINARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM T. AQUATICUS BOUND TO A PRIMER/TEMPLATE DNA 4n56 
4N56 4n5s 
4N5S 4o0i 
4O0I 4uqg 
4UQG 4x0p 
4X0P 4x0q 
4X0Q 4xiu 
4XIU 4xvi 
4XVI 4xvk 
4XVK 4xvl 
4XVL 4xvm 
4XVM 4yfu 
4YFU 4ztu 
4ZTU 4ztz 
4ZTZ 5c51 
5C51 5c52 
5C52 5c53 
5C53 5dkt 
5DKT 5dku 
5DKU 5ktq 
LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP - Links (links to other resources describing this domain)
-
PROSITE DNA_POLYMERASE_A INTERPRO IPR001098 PFAM DNA_pol_A

